Make a scatterplot
create.scatterplot.RdTakes a data.frame and creates a scatterplot
Usage
create.scatterplot(
formula,
data,
filename = NULL,
groups = NULL,
main = NULL,
main.just = 'center',
main.x = 0.5,
main.y = 0.5,
main.cex = 3,
xlab.label = tail(sub('~', '', formula[-2]), 1),
ylab.label = tail(sub('~', '', formula[-3]), 1),
xlab.cex = 2,
ylab.cex = 2,
xlab.col = 'black',
ylab.col = 'black',
xlab.top.label = NULL,
xlab.top.cex = 2,
xlab.top.col = 'black',
xlab.top.just = 'center',
xlab.top.x = 0.5,
xlab.top.y = 0,
xlimits = NULL,
ylimits = NULL,
xat = TRUE,
yat = TRUE,
xaxis.lab = NA,
yaxis.lab = NA,
xaxis.log = FALSE,
yaxis.log = FALSE,
xaxis.cex = 1.5,
yaxis.cex = 1.5,
xaxis.rot = 0,
yaxis.rot = 0,
xaxis.fontface = 'bold',
yaxis.fontface = 'bold',
xaxis.col = 'black',
yaxis.col = 'black',
xaxis.tck = c(1,1),
yaxis.tck = c(1,1),
add.grid = FALSE,
xgrid.at = xat,
ygrid.at = yat,
grid.colour = NULL,
horizontal = FALSE,
type = 'p',
cex = 0.75,
pch = 19,
col = 'black',
col.border = 'black',
lwd = 1,
lty = 1,
alpha = 1,
axes.lwd = 1,
strip.col = 'white',
strip.cex = 1,
strip.fontface = 'bold',
y.error.up = NULL,
y.error.down = y.error.up,
x.error.right = NULL,
x.error.left = x.error.right,
y.error.bar.col = 'black',
x.error.bar.col = y.error.bar.col,
error.whisker.angle = 90,
error.bar.lwd = 1,
error.bar.length = 0.1,
key = list(text = list(lab = c(''))),
legend = NULL,
top.padding = 0.1,
bottom.padding = 0.7,
right.padding = 0.1,
left.padding = 0.5,
key.top = 0.1,
key.left.padding = 0,
ylab.axis.padding = 1,
axis.key.padding = 1,
layout = NULL,
as.table = FALSE,
x.spacing = 0,
y.spacing = 0,
x.relation = 'same',
y.relation = 'same',
add.axes = FALSE,
axes.lty = 'dashed',
add.xyline = FALSE,
xyline.col = 'black',
xyline.lwd = 1,
xyline.lty = 1,
abline.h = NULL,
abline.v = NULL,
abline.col = 'black',
abline.lwd = 1,
abline.lty = 1,
add.curves = FALSE,
curves.exprs = NULL,
curves.from = min(data, na.rm = TRUE),
curves.to = max(data, na.rm = TRUE),
curves.col = 'black',
curves.lwd = 2,
curves.lty = 1,
add.rectangle = FALSE,
xleft.rectangle = NULL,
ybottom.rectangle = NULL,
xright.rectangle = NULL,
ytop.rectangle = NULL,
col.rectangle = 'transparent',
alpha.rectangle = 1,
add.points = FALSE,
points.x = NULL,
points.y = NULL,
points.pch = 19,
points.col = 'black',
points.col.border = 'black',
points.cex = 1,
add.line.segments = FALSE,
line.start = NULL,
line.end = NULL,
line.col = 'black',
line.lwd = 1,
add.text = FALSE,
text.labels = NULL,
text.x = NULL,
text.y = NULL,
text.col = 'black',
text.cex = 1,
text.fontface = 'bold',
text.guess.labels = FALSE,
text.guess.skip.labels = TRUE,
text.guess.ignore.radius = FALSE,
text.guess.ignore.rectangle = FALSE,
text.guess.radius.factor = 1,
text.guess.buffer.factor = 1,
text.guess.label.position = NULL,
height = 6,
width = 6,
size.units = 'in',
resolution = 1600,
enable.warnings = FALSE,
description = 'Created with BoutrosLab.plotting.general',
style = 'BoutrosLab',
preload.default = 'custom',
group.specific.colouring = TRUE,
use.legacy.settings = FALSE,
inside.legend.auto = FALSE,
regions.labels = c(),
regions.start = c(),
regions.stop = c(),
regions.color = c("red"),
regions.cex = 1,
regions.alpha = 1,
lollipop.bar.y = NULL,
lollipop.bar.color = "gray",
...
);Arguments
- formula
The formula used to extract the x & y components from the data-frame. Transforming data within formula is not compatible with automatic scaling with `xat` or `yat`.
- data
The data-frame to plot
- filename
Filename for tiff output, or if NULL returns the trellis object itself
- groups
The grouping variable in the data-frame
- main
The main title for the plot (space is reclaimed if NULL)
- main.just
The justification of the main title for the plot, default is centered
- main.x
The x location of the main title, deault is 0.5
- main.y
The y location of the main title, default is 0.5
- main.cex
Size of text for main plot title
- xlab.label
x-axis label
- ylab.label
y-axis label
- xlab.cex
Size of x-axis label, defaults to 3
- ylab.cex
Size of y-axis label, defaults to 3
- xlab.col
Colour of the x-axis label, defaults to “black”
- ylab.col
Colour of the y-axis label, defaults to “black”
- xlab.top.label
The label for the top x-axis
- xlab.top.cex
Size of top x-axis label
- xlab.top.col
Colour of the top x-axis label
- xlab.top.just
Justification of the top x-axis label, defaults to centered
- xlab.top.x
The x location of the top x-axis label
- xlab.top.y
The y location of the top y-axis label
- xlimits
Two-element vector giving the x-axis limits, defaults to automatic
- ylimits
Two-element vector giving the y-axis limits, defaults to automatic
- xat
Accepts a vector listing where x-axis ticks should be drawn or if automatic scaling is desired, one of three strings: “auto”, “auto.linear” or “auto.log”. Automatic scaling fixes x-axis tick locations, labels, and data values dependent given data. “auto” will determine whether linear or logarithmic scaling fits the given data best, “auto.linear” or “auto.log” will force data to be scaled linearly or logarithmically respectively. Defaults to lattice automatic (TRUE). For more details see `auto.axis()`.
- yat
Accepts a vector listing where y-axis ticks should be drawn or if automatic scaling is desired, one of three strings: “auto”, “auto.linear” or “auto.log”. Automatic scaling fixes y-axis tick locations, labels, and data values dependent given data. “auto” will determine whether linear or logarithmic scaling fits the given data best, “auto.linear” or “auto.log” will force data to be scaled linearly or logarithmically respectively. Defaults to lattice automatic (TRUE). For more details see `auto.axis()`.
- xaxis.lab
Vector listing x-axis tick labels, defaults to automatic (TRUE). Using automatic scaling with xat will overwrite user input. Set to NULL to remove x-axis labels.
- yaxis.lab
Vector listing y-axis tick labels, defaults to automatic (TRUE). Using automatic scaling with yat will overwrite user input. Set to NULL to remove y-axis labels.
- xaxis.log
Logical indicating whether x-variable should be in logarithmic scale (and what base if numeric)
- yaxis.log
Logical indicating whether y-variable should be in logarithmic scale (and what base if numeric)
- xaxis.cex
Size of x-axis scales, defaults to 2
- yaxis.cex
Size of y-axis scales, defaults to 2
- xaxis.rot
Counterclockwise rotation of text in x-axis scales in degrees, defaults to 0
- yaxis.rot
Counterclockwise rotation of text in y-axis scales in degrees, defaults to 0
- xaxis.fontface
Fontface for the x-axis scales
- yaxis.fontface
Fontface for the y-axis scales
- xaxis.col
Colour of the x-axis tick labels, defaults to “black”
- yaxis.col
Colour of the y-axis tick labels, defaults to “black”
- xaxis.tck
Specifies the length of the tick mark, defaults to 1 for both top and bottom axes
- yaxis.tck
Specifies the length of the tick mark, defaults to 1 for both top and bottom axes
- add.grid
Logical stating wheter or not the grid should be drawn on the plot
- xgrid.at
Vector listing where the x-axis grid lines should be drawn, defaults to xat
- ygrid.at
Vector listing where the y-axis grid lines should be drawn, defaults to yat
- grid.colour
ability to set individual grid line colours
- horizontal
xyplot-specific function that allows you to change if type='h' draws lines to the vertical or horizontal axis
- type
Accepts character vector of one or more elements defining how x and y are to be plotted. Accepted elements include: "p" to draw points, "l" to connect points with lines, "h" to draw vertical or horizonal line segments from the points to the origin, "s" or "S" to plot as a step curve, "g" to add a grid, and "r" to add a linear regression line. For more options and detail see "type" parameter in "xyplot" documentation.
- cex
Character expansion for plotting symbol
- pch
Plotting character
- col
Point/line colour
- col.border
Colour of border when points pch >= 21. Defaults to “black”
- lwd
Specifies line width, defaults to 1
- lty
Specifies line style, defaults to 1 (solid)
- alpha
Specifies line transparency, defaults to 1 (opaque)
- axes.lwd
Thickness of width of axes lines
- strip.col
Strip background colour, defaults to “white”
- strip.cex
Strip title character expansion
- strip.fontface
Strip title fontface, defaults to bold
- y.error.up
upward error vector. Defaults to NULL. When y.error.up is NULL, vertical error bar is not drawn
- y.error.down
Downward error vector. Defaults to y.error.down to show symmetric error bars
- x.error.right
Rightward error vector. Defaults to NULL. When x.error.right is NULL, horizontal error bar is not drawn
- x.error.left
Leftward error vector. Defaults to x.error.right to show symmetric error bars
- y.error.bar.col
Colour of vertical error bar. Defaults to “black”
- x.error.bar.col
Colour of horizontal error bar. Defaults to “black”
- error.whisker.angle
Angle of the whisker drawn on error bar. Defaults to 90 degree
- error.bar.lwd
Error bar line width. Defaults to 1
- error.bar.length
Length of the error bar whiskers. Defaults to 0.1
- key
A list giving the key (legend). The default suppresses drawing
- legend
Add a legend to the plot. Helpful for adding multiple keys and adding keys to the margins of the plot. See xyplot.
- top.padding
A number specifying the distance to the top margin, defaults to 0.1
- bottom.padding
A number specifying the distance to the bottom margin, defaults to 0.7
- right.padding
A number specifying the distance to the right margin, defaults to 0.1
- left.padding
A number specifying the distance to the left margin, defaults to 0.5
- key.top
A number specifying the distance at top of key, defaults to 0.1
- key.left.padding
Amount of padding to go onto any legend on the left
- ylab.axis.padding
A number specifying the distance of ylabel to the y-axis, defaults to 1
- axis.key.padding
A number specifying the distance from the y-axis to the key, defaults to 1
- layout
A vector specifying the number of columns, rows (e.g., c(2,1). Default is NULL; see lattice::xyplot for more details
- as.table
Specifies panel drawing order, default is FALSE which draws panels from bottom left corner, moving right then up. Set to TRUE to draw from top left corner, moving right then down
- x.spacing
A number specifying the distance between panels along the x-axis, defaults to 0
- y.spacing
A number specifying the distance between panels along the y-axis, defaults to 0
- x.relation
Allows x-axis scales to vary if set to “free”, defaults to “same”
- y.relation
Allows y-axis scales to vary if set to “free”, defaults to “same”
- add.axes
Allow axis lines to be turned on or off, default is FALSE
- axes.lty
Specifies axis line style, defaults to “dashed”
- add.xyline
Allow y=x line to be drawn, default is FALSE
- xyline.col
y=x line colour, defaults to black
- xyline.lwd
Specifies y=x line width, defaults to 1
- xyline.lty
Specifies y=x line style, defaults to 1 (solid)
- abline.h
Allow horizontal line to be drawn, default to NULL
- abline.v
Allow vertical line to be drawn, default to NULL
- abline.col
Horizontal line colour, defaults to black
- abline.lwd
Specifies horizontal line width, defaults to 1
- abline.lty
Specifies horizontal line style, defaults to 1 (solid)
- add.curves
Allow curves to drawn, default is FALSE
- curves.exprs
A list of functions, expressions, or calls using “x” as a variable that specify the curves to be drawn
- curves.from
Specifies the x co-ordinates at which the start of each curve should be drawn, defaults to drawing the curves to the left edge of the plotting region
- curves.to
Specifies the x co-ordinates at which the end of each curve should be drawn, defaults to drawing the curves to the right edge of the plotting region
- curves.col
Specifies colours of curves, default is black for each curve
- curves.lwd
Specifies width of curves, default is 1 for each curve
- curves.lty
Specifies type of curves, default is 1 (solid) for each curve
- add.rectangle
Allow a rectangle to be drawn, default is FALSE
- xleft.rectangle
Specifies the left x ooordinate of the rectangle to be drawn
- ybottom.rectangle
Specifies the bottom y coordinate of the rectangle to be drawn
- xright.rectangle
Specifies the right x coordinate of the rectangle to be drawn
- ytop.rectangle
Specifies the top y coordinate of the rectangle to be drawn
- col.rectangle
Specifies the colour to fill rectangle's area
- alpha.rectangle
Specifies the colour bias of the rectangle to be drawn
- add.points
Allow additional points to be drawn, default is FALSE
- points.x
The x co-ordinates where additional points should be drawn
- points.y
The y co-ordinates where additional points should be drawn
- points.pch
The plotting character for additional points
- points.col
The colour of additional points
- points.col.border
Colour of the border of additional points if points.pch >= 21. Defaults to black
- points.cex
The size of additional points
- add.line.segments
Allow additional line segments to be drawn, default is FALSE
- line.start
The y co-ordinates where additional line segments should start
- line.end
The y co-ordinates where additional line segments should end
- line.col
The colour of additional line segments, default is black
- line.lwd
The line width of additional line segments, default is 1
- add.text
Allow additional text to be drawn, default is FALSE
- text.labels
Labels for additional text
- text.x
The x co-ordinates where additional text should be placed
- text.y
The y co-ordinates where additional text should be placed
- text.col
The colour of additional text
- text.cex
The size of additional text
- text.fontface
The fontface for additional text
- text.guess.labels
Allows automatic labeling by considering values in text.x and text.y as a data point to be labelled, default is FALSE
- text.guess.skip.labels
Provides an option to disregard automatic labelling algorithm if no space is available around a data point, thus forcing labelling if a collision is likely, default is TRUE
- text.guess.ignore.radius
Allows the automatic labeling algorithm to ignore the radius space of a data point, useful to label a cluster of data points with a single text box, default is FALSE
- text.guess.ignore.rectangle
Allows the atuomatic labeling algorithm to ignore the rectangle space of multiple potential label positions, default is FALSE
- text.guess.radius.factor
A numeric value to factor the radius value to alter distance from the label and the data point
- text.guess.buffer.factor
A numeric value to factor the buffer value to alter the space which is used to consider if data.points are potentially going to collide
- text.guess.label.position
A numeric value between 0 and 360 to specify the percise angle of a text box center and the positive x-axis. Angles move counter-clockwise beginning at the positive x axis
- height
Figure height, defaults to 6 inches
- width
Figure width, defaults to 6 inches
- size.units
Figure units, defaults to inches
- resolution
Figure resolution in dpi, defaults to 1600
- enable.warnings
Print warnings if set to TRUE, defaults to FALSE
- description
Short description of image/plot; default NULL
- style
defaults to “BoutrosLab”, also accepts “Nature”, which changes parameters according to Nature formatting requirements
- preload.default
ability to set multiple sets of diffrent defaults depending on publication needs
- group.specific.colouring
Variable to specify if group specific multi colouring for error bars is enforced
- use.legacy.settings
boolean to set wheter or not to use legacy mode settings (font)
- inside.legend.auto
boolean specifying whether or not to use the automatic inside legend function
- regions.labels
Labels for each of the regions on the lollipop plots bars
- regions.start
start x value of each of the regions
- regions.stop
stop value for each of the regions
- regions.color
color of each of the regions
- regions.cex
size of the text of each of the regions
- regions.alpha
alpha of each of the regions
- lollipop.bar.y
y location of top of the lollipop plot bar – defaults to right above the bottom y axis
- lollipop.bar.color
color of the lollipop plot bar
- ...
Additional arguments to be passed to xyplot
Value
If filename is NULL then returns the trellis object, otherwise creates a plot and returns a 0/1 success code.
Warning
If this function is called without capturing the return value, or specifying a filename, it may crash while trying to draw the histogram. In particular, if a script that uses such a call of create histogram is called by reading the script in from the command line, it will fail badly, with an error message about unavailable fonts:
Error in grid.Call.graphics("L_text", as.graphicsAnnot(x$label), x$x, )
Invalid font type
Calls: print ... drawDetails.text -> grid.Call.graphics -> .Call.graphics
Examples
set.seed(12345);
simple.data <- data.frame(
x = rnorm(800),
y = rnorm(800)
);
create.scatterplot(
# # filename = tempfile(pattern = 'Scatterplot_Simple', fileext = '.tiff'),
formula = y ~ x,
data = simple.data,
resolution = 50
);
 scatter.data <- data.frame(
sample.one = microarray[1:800,1],
sample.two = microarray[1:800,2],
chr = microarray$Chr[1:800]
);
# Minimal Input
create.scatterplot(
# filename = tempfile(pattern = 'Scatterplot_Minimal_Input', fileext = '.tiff'),
formula = sample.two ~ sample.one,
data = scatter.data,
main = 'Minimal Input',
description = 'Scatter plot created by BoutrosLab.plotting.general',
resolution = 50
);
scatter.data <- data.frame(
sample.one = microarray[1:800,1],
sample.two = microarray[1:800,2],
chr = microarray$Chr[1:800]
);
# Minimal Input
create.scatterplot(
# filename = tempfile(pattern = 'Scatterplot_Minimal_Input', fileext = '.tiff'),
formula = sample.two ~ sample.one,
data = scatter.data,
main = 'Minimal Input',
description = 'Scatter plot created by BoutrosLab.plotting.general',
resolution = 50
);
 # Axes & Labels
create.scatterplot(
# filename = tempfile(pattern = 'Scatterplot_Axes_Labels', fileext = '.tiff'),
formula = sample.two ~ sample.one,
data = scatter.data,
main = 'Axes & Labels',
# Axes and labels
xlab.label = colnames(microarray[1]),
ylab.label = colnames(microarray[2]),
xat = seq(0, 16, 2),
yat = seq(0, 16, 2),
xlimits = c(0, 15),
ylimits = c(0, 15),
xaxis.cex = 1,
yaxis.cex = 1,
xaxis.fontface = 1,
yaxis.fontface = 1,
xlab.cex = 1.5,
ylab.cex = 1.5,
description = 'Scatter plot created by BoutrosLab.plotting.general',
resolution = 50
);
# Axes & Labels
create.scatterplot(
# filename = tempfile(pattern = 'Scatterplot_Axes_Labels', fileext = '.tiff'),
formula = sample.two ~ sample.one,
data = scatter.data,
main = 'Axes & Labels',
# Axes and labels
xlab.label = colnames(microarray[1]),
ylab.label = colnames(microarray[2]),
xat = seq(0, 16, 2),
yat = seq(0, 16, 2),
xlimits = c(0, 15),
ylimits = c(0, 15),
xaxis.cex = 1,
yaxis.cex = 1,
xaxis.fontface = 1,
yaxis.fontface = 1,
xlab.cex = 1.5,
ylab.cex = 1.5,
description = 'Scatter plot created by BoutrosLab.plotting.general',
resolution = 50
);
 # Log-Scaled Axis
log.data <- data.frame(
x = rnorm(800),
y = 10 ** rnorm(800, mean = 5, sd = 2)
);
create.scatterplot(
formula = y ~ x,
data = log.data,
# Log base 10 scale y-axis
yat = 'auto.log',
main = 'Log Scaled',
description = 'Scatter created by BoutrosLab.plotting.general',
resolution = 50
);
# Log-Scaled Axis
log.data <- data.frame(
x = rnorm(800),
y = 10 ** rnorm(800, mean = 5, sd = 2)
);
create.scatterplot(
formula = y ~ x,
data = log.data,
# Log base 10 scale y-axis
yat = 'auto.log',
main = 'Log Scaled',
description = 'Scatter created by BoutrosLab.plotting.general',
resolution = 50
);
 # Colour & Plotting Character
create.scatterplot(
# filename = tempfile(pattern = 'Scatterplot_Colour_Pch', fileext = '.tiff'),
formula = sample.two ~ sample.one,
data = scatter.data,
main = 'Colour & Pch',
xlab.label = colnames(microarray[1]),
ylab.label = colnames(microarray[2]),
xat = seq(0, 16, 2),
yat = seq(0, 16, 2),
xlimits = c(0, 15),
ylimits = c(0, 15),
xaxis.cex = 1,
yaxis.cex = 1,
xaxis.fontface = 1,
yaxis.fontface = 1,
xlab.cex = 1.5,
ylab.cex = 1.5,
# setting the colour
col = default.colours(2)[2],
# setting the plotting character type & size
pch = 21,
cex = 1.5,
description = 'Scatter plot created by BoutrosLab.plotting.general',
resolution = 100
);
# Colour & Plotting Character
create.scatterplot(
# filename = tempfile(pattern = 'Scatterplot_Colour_Pch', fileext = '.tiff'),
formula = sample.two ~ sample.one,
data = scatter.data,
main = 'Colour & Pch',
xlab.label = colnames(microarray[1]),
ylab.label = colnames(microarray[2]),
xat = seq(0, 16, 2),
yat = seq(0, 16, 2),
xlimits = c(0, 15),
ylimits = c(0, 15),
xaxis.cex = 1,
yaxis.cex = 1,
xaxis.fontface = 1,
yaxis.fontface = 1,
xlab.cex = 1.5,
ylab.cex = 1.5,
# setting the colour
col = default.colours(2)[2],
# setting the plotting character type & size
pch = 21,
cex = 1.5,
description = 'Scatter plot created by BoutrosLab.plotting.general',
resolution = 100
);
 # Colour depth
# create colour scheme to illustrate adding a colourkey
chr.palette <- colour.gradient(default.colours(2)[2], 800);
create.scatterplot(
# filename = tempfile(pattern = 'Scatterplot_Colour_Depth', fileext = '.tiff'),
formula = sample.two ~ sample.one,
data = scatter.data,
main = 'Colour Depth',
xlab.label = colnames(microarray[1]),
ylab.label = colnames(microarray[2]),
xat = seq(0, 16, 2),
yat = seq(0, 16, 2),
xlimits = c(0, 15),
ylimits = c(0, 15),
xaxis.cex = 1,
yaxis.cex = 1,
xaxis.fontface = 1,
yaxis.fontface = 1,
xlab.cex = 1.5,
ylab.cex = 1.5,
# setting the colour
col = chr.palette,
# setting the plotting character type & size
pch = 19,
cex = 1,
# adding key for colours
key.top = 1.5,
legend = list(
bottom = list(
fun = draw.colorkey,
args = list(
key = list(
col = chr.palette,
at = 1:800,
tick.number = 3,
space = 'bottom',
size = 1,
width = 1.25,
height = 1,
labels = list(
labels = 1:3,
cex = 1,
at = c(1, which(scatter.data$chr == 2)[1], which(scatter.data$chr == 3)[1])
)
)
)
)
),
description = 'Scatter plot created by BoutrosLab.plotting.general',
resolution = 100
);
# Colour depth
# create colour scheme to illustrate adding a colourkey
chr.palette <- colour.gradient(default.colours(2)[2], 800);
create.scatterplot(
# filename = tempfile(pattern = 'Scatterplot_Colour_Depth', fileext = '.tiff'),
formula = sample.two ~ sample.one,
data = scatter.data,
main = 'Colour Depth',
xlab.label = colnames(microarray[1]),
ylab.label = colnames(microarray[2]),
xat = seq(0, 16, 2),
yat = seq(0, 16, 2),
xlimits = c(0, 15),
ylimits = c(0, 15),
xaxis.cex = 1,
yaxis.cex = 1,
xaxis.fontface = 1,
yaxis.fontface = 1,
xlab.cex = 1.5,
ylab.cex = 1.5,
# setting the colour
col = chr.palette,
# setting the plotting character type & size
pch = 19,
cex = 1,
# adding key for colours
key.top = 1.5,
legend = list(
bottom = list(
fun = draw.colorkey,
args = list(
key = list(
col = chr.palette,
at = 1:800,
tick.number = 3,
space = 'bottom',
size = 1,
width = 1.25,
height = 1,
labels = list(
labels = 1:3,
cex = 1,
at = c(1, which(scatter.data$chr == 2)[1], which(scatter.data$chr == 3)[1])
)
)
)
)
),
description = 'Scatter plot created by BoutrosLab.plotting.general',
resolution = 100
);
 # Groups & Legend
create.scatterplot(
# filename = tempfile(pattern = 'Scatterplot_Groups_Legend', fileext = '.tiff'),
formula = sample.two ~ sample.one,
data = scatter.data,
main = 'Groups & Legend',
# using arbitrary groups for the sake of illustration
groups = scatter.data$chr,
xlab.label = colnames(microarray[1]),
ylab.label = colnames(microarray[2]),
xat = seq(0, 16, 2),
yat = seq(0, 16, 2),
xlimits = c(0, 15),
ylimits = c(0, 15),
xaxis.cex = 1,
yaxis.cex = 1,
xaxis.fontface = 1,
yaxis.fontface = 1,
xlab.cex = 1.5,
ylab.cex = 1.5,
col = default.colours(3),
# Adding legend for groups
key = list(
text = list(
lab = c('1','2','3'),
cex = 1,
col = 'black'
),
points = list(
pch = 19,
col = default.colours(3),
cex = 1
),
x = 0.04,
y = 0.95,
padding.text = 2
),
description = 'Scatter plot created by BoutrosLab.plotting.general',
resolution = 100
);
# Groups & Legend
create.scatterplot(
# filename = tempfile(pattern = 'Scatterplot_Groups_Legend', fileext = '.tiff'),
formula = sample.two ~ sample.one,
data = scatter.data,
main = 'Groups & Legend',
# using arbitrary groups for the sake of illustration
groups = scatter.data$chr,
xlab.label = colnames(microarray[1]),
ylab.label = colnames(microarray[2]),
xat = seq(0, 16, 2),
yat = seq(0, 16, 2),
xlimits = c(0, 15),
ylimits = c(0, 15),
xaxis.cex = 1,
yaxis.cex = 1,
xaxis.fontface = 1,
yaxis.fontface = 1,
xlab.cex = 1.5,
ylab.cex = 1.5,
col = default.colours(3),
# Adding legend for groups
key = list(
text = list(
lab = c('1','2','3'),
cex = 1,
col = 'black'
),
points = list(
pch = 19,
col = default.colours(3),
cex = 1
),
x = 0.04,
y = 0.95,
padding.text = 2
),
description = 'Scatter plot created by BoutrosLab.plotting.general',
resolution = 100
);
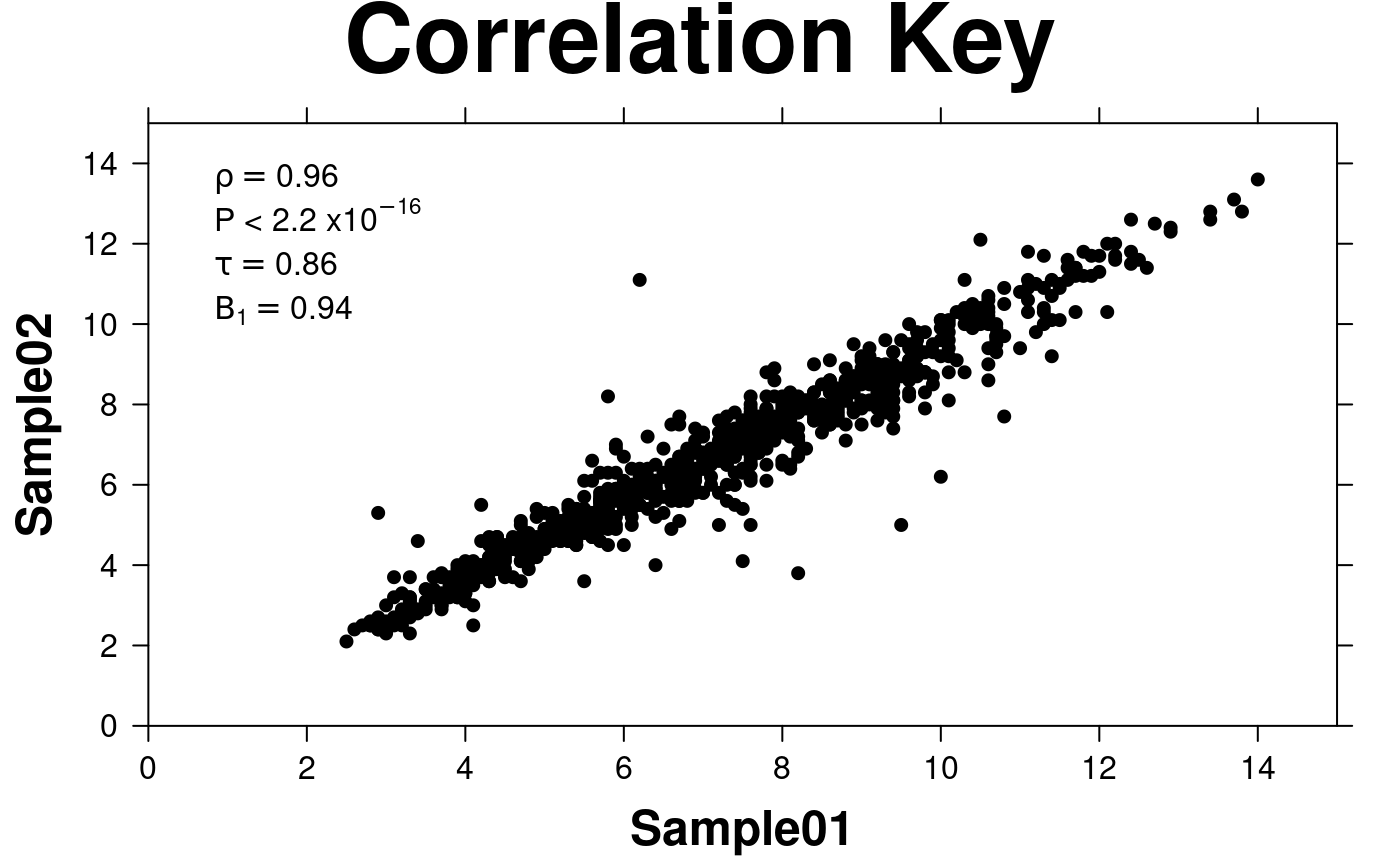
 # Correlation Key
create.scatterplot(
# filename = tempfile(pattern = 'Scatterplot_Correlation_Key', fileext = '.tiff'),
formula = sample.two ~ sample.one,
data = scatter.data,
main = 'Correlation Key',
xlab.label = colnames(microarray[1]),
ylab.label = colnames(microarray[2]),
xat = seq(0, 16, 2),
yat = seq(0, 16, 2),
xlimits = c(0, 15),
ylimits = c(0, 15),
xaxis.cex = 1,
yaxis.cex = 1,
xaxis.fontface = 1,
yaxis.fontface = 1,
xlab.cex = 1.5,
ylab.cex = 1.5,
col = 'black',
pch = 21,
# Adding correlation key
legend = list(
inside = list(
fun = draw.key,
args = list(
key = get.corr.key(
x = scatter.data$sample.one,
y = scatter.data$sample.two,
label.items = c('spearman','spearman.p','kendall','beta1'),
alpha.background = 0,
key.cex = 1
)
),
x = 0.04,
y = 0.95,
corner = c(0,1)
)
),
description = 'Scatter plot created by BoutrosLab.plotting.general',
resolution = 100
);
#> Warning: Cannot compute exact p-value with ties
# Correlation Key
create.scatterplot(
# filename = tempfile(pattern = 'Scatterplot_Correlation_Key', fileext = '.tiff'),
formula = sample.two ~ sample.one,
data = scatter.data,
main = 'Correlation Key',
xlab.label = colnames(microarray[1]),
ylab.label = colnames(microarray[2]),
xat = seq(0, 16, 2),
yat = seq(0, 16, 2),
xlimits = c(0, 15),
ylimits = c(0, 15),
xaxis.cex = 1,
yaxis.cex = 1,
xaxis.fontface = 1,
yaxis.fontface = 1,
xlab.cex = 1.5,
ylab.cex = 1.5,
col = 'black',
pch = 21,
# Adding correlation key
legend = list(
inside = list(
fun = draw.key,
args = list(
key = get.corr.key(
x = scatter.data$sample.one,
y = scatter.data$sample.two,
label.items = c('spearman','spearman.p','kendall','beta1'),
alpha.background = 0,
key.cex = 1
)
),
x = 0.04,
y = 0.95,
corner = c(0,1)
)
),
description = 'Scatter plot created by BoutrosLab.plotting.general',
resolution = 100
);
#> Warning: Cannot compute exact p-value with ties
 # \donttest{
# Panel Organization
create.scatterplot(
# filename = tempfile(pattern = 'Scatterplot_Panel_numeric_conditional', fileext = '.tiff'),
formula = sample.two ~ sample.one | chr,
data = scatter.data,
main = 'Panel',
xlab.label = colnames(microarray[1]),
ylab.label = colnames(microarray[2]),
xat = seq(0, 16, 2),
yat = seq(0, 16, 2),
xlimits = c(0, 15),
ylimits = c(0, 15),
xaxis.cex = 1,
yaxis.cex = 1,
xaxis.fontface = 1,
yaxis.fontface = 1,
xlab.cex = 1.5,
ylab.cex = 1.5,
pch = 21,
col = 'black',
fill = 'transparent',
# set up panel layout
layout = c(1,3),
yrelation = 'free',
description = 'Scatter plot created by BoutrosLab.plotting.general',
resolution = 100
);
#> Warning: Numeric values detected for conditional variable. If text labels are desired, please convert conditional variable to character.
# \donttest{
# Panel Organization
create.scatterplot(
# filename = tempfile(pattern = 'Scatterplot_Panel_numeric_conditional', fileext = '.tiff'),
formula = sample.two ~ sample.one | chr,
data = scatter.data,
main = 'Panel',
xlab.label = colnames(microarray[1]),
ylab.label = colnames(microarray[2]),
xat = seq(0, 16, 2),
yat = seq(0, 16, 2),
xlimits = c(0, 15),
ylimits = c(0, 15),
xaxis.cex = 1,
yaxis.cex = 1,
xaxis.fontface = 1,
yaxis.fontface = 1,
xlab.cex = 1.5,
ylab.cex = 1.5,
pch = 21,
col = 'black',
fill = 'transparent',
# set up panel layout
layout = c(1,3),
yrelation = 'free',
description = 'Scatter plot created by BoutrosLab.plotting.general',
resolution = 100
);
#> Warning: Numeric values detected for conditional variable. If text labels are desired, please convert conditional variable to character.
 scatter.data$chromosome <- as.character(scatter.data$chr);
create.scatterplot(
# filename = tempfile(pattern = 'Scatterplot_Panel_character_conditional', fileext = '.tiff'),
formula = sample.two ~ sample.one | chromosome,
data = scatter.data,
main = 'Panel',
xlab.label = colnames(microarray[1]),
ylab.label = colnames(microarray[2]),
xat = seq(0, 16, 2),
yat = seq(0, 16, 2),
xlimits = c(0, 15),
ylimits = c(0, 15),
xaxis.cex = 1,
yaxis.cex = 1,
xaxis.fontface = 1,
yaxis.fontface = 1,
xlab.cex = 1.5,
ylab.cex = 1.5,
pch = 21,
col = 'black',
fill = 'transparent',
# set up panel layout
layout = c(1,3),
yrelation = 'free',
description = 'Scatter plot created by BoutrosLab.plotting.general',
resolution = 100
);
scatter.data$chromosome <- as.character(scatter.data$chr);
create.scatterplot(
# filename = tempfile(pattern = 'Scatterplot_Panel_character_conditional', fileext = '.tiff'),
formula = sample.two ~ sample.one | chromosome,
data = scatter.data,
main = 'Panel',
xlab.label = colnames(microarray[1]),
ylab.label = colnames(microarray[2]),
xat = seq(0, 16, 2),
yat = seq(0, 16, 2),
xlimits = c(0, 15),
ylimits = c(0, 15),
xaxis.cex = 1,
yaxis.cex = 1,
xaxis.fontface = 1,
yaxis.fontface = 1,
xlab.cex = 1.5,
ylab.cex = 1.5,
pch = 21,
col = 'black',
fill = 'transparent',
# set up panel layout
layout = c(1,3),
yrelation = 'free',
description = 'Scatter plot created by BoutrosLab.plotting.general',
resolution = 100
);
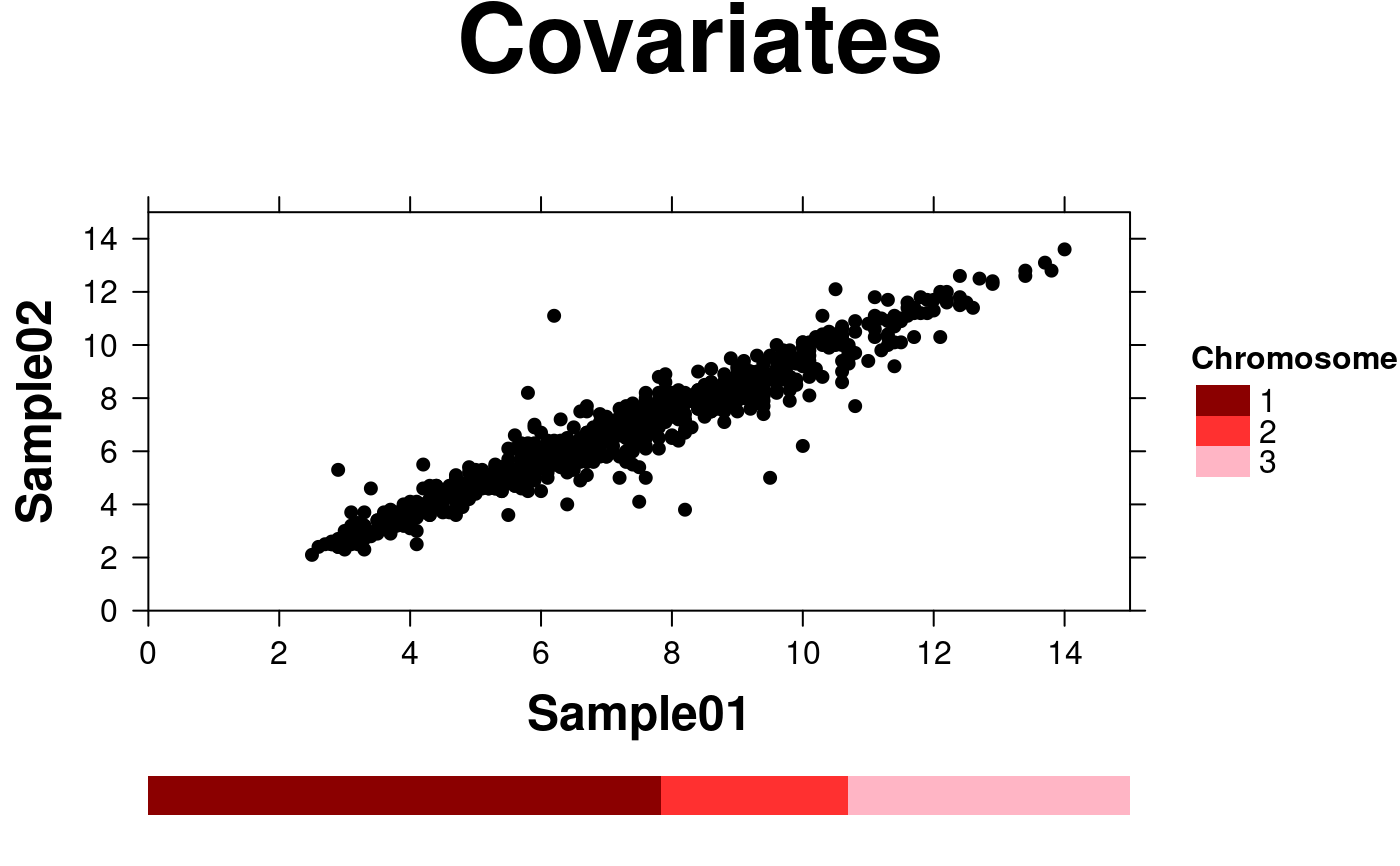
 # Covariates
cov.colours <- as.character(microarray$Chr[1:800]);
cov.colours[cov.colours == '1'] <- default.colours(3, palette.type = 'chromosomes')[1];
cov.colours[cov.colours == '2'] <- default.colours(3, palette.type = 'chromosomes')[2];
cov.colours[cov.colours == '3'] <- default.colours(3, palette.type = 'chromosomes')[3];
cov <- list(
rect = list(
col = 'transparent',
fill = cov.colours
)
);
cov.grob <- covariates.grob(
covariates = cov,
ord = c(1:length(cov.colours)),
side = 'top',
size = 1
);
cov.legend <- list(
legend = list(
colours = default.colours(3, palette.type = 'chromosomes'),
labels = c('1', '2', '3'),
title = 'Chromosome',
border = 'transparent'
)
);
cov.legend.grob <- legend.grob(
legends = cov.legend
);
create.scatterplot(
# filename = tempfile(pattern = 'Scatterplot_Covariates', fileext = '.tiff'),
formula = sample.two ~ sample.one,
data = scatter.data,
main = 'Covariates',
xlab.label = colnames(microarray[1]),
ylab.label = colnames(microarray[2]),
xat = seq(0, 16, 2),
yat = seq(0, 16, 2),
xlimits = c(0, 15),
ylimits = c(0, 15),
xaxis.cex = 1,
yaxis.cex = 1,
xaxis.fontface = 1,
yaxis.fontface = 1,
xlab.cex = 1.5,
ylab.cex = 1.5,
pch = 21,
col = 'black',
fill = 'transparent',
# Adding covariate & legend
legend = list(
bottom = list(fun = cov.grob),
right = list(fun = cov.legend.grob)
),
# Ensuring sufficient spacing for covariate
key.top = 3,
description = 'Scatter plot created by BoutrosLab.plotting.general',
resolution = 200
);
# Covariates
cov.colours <- as.character(microarray$Chr[1:800]);
cov.colours[cov.colours == '1'] <- default.colours(3, palette.type = 'chromosomes')[1];
cov.colours[cov.colours == '2'] <- default.colours(3, palette.type = 'chromosomes')[2];
cov.colours[cov.colours == '3'] <- default.colours(3, palette.type = 'chromosomes')[3];
cov <- list(
rect = list(
col = 'transparent',
fill = cov.colours
)
);
cov.grob <- covariates.grob(
covariates = cov,
ord = c(1:length(cov.colours)),
side = 'top',
size = 1
);
cov.legend <- list(
legend = list(
colours = default.colours(3, palette.type = 'chromosomes'),
labels = c('1', '2', '3'),
title = 'Chromosome',
border = 'transparent'
)
);
cov.legend.grob <- legend.grob(
legends = cov.legend
);
create.scatterplot(
# filename = tempfile(pattern = 'Scatterplot_Covariates', fileext = '.tiff'),
formula = sample.two ~ sample.one,
data = scatter.data,
main = 'Covariates',
xlab.label = colnames(microarray[1]),
ylab.label = colnames(microarray[2]),
xat = seq(0, 16, 2),
yat = seq(0, 16, 2),
xlimits = c(0, 15),
ylimits = c(0, 15),
xaxis.cex = 1,
yaxis.cex = 1,
xaxis.fontface = 1,
yaxis.fontface = 1,
xlab.cex = 1.5,
ylab.cex = 1.5,
pch = 21,
col = 'black',
fill = 'transparent',
# Adding covariate & legend
legend = list(
bottom = list(fun = cov.grob),
right = list(fun = cov.legend.grob)
),
# Ensuring sufficient spacing for covariate
key.top = 3,
description = 'Scatter plot created by BoutrosLab.plotting.general',
resolution = 200
);
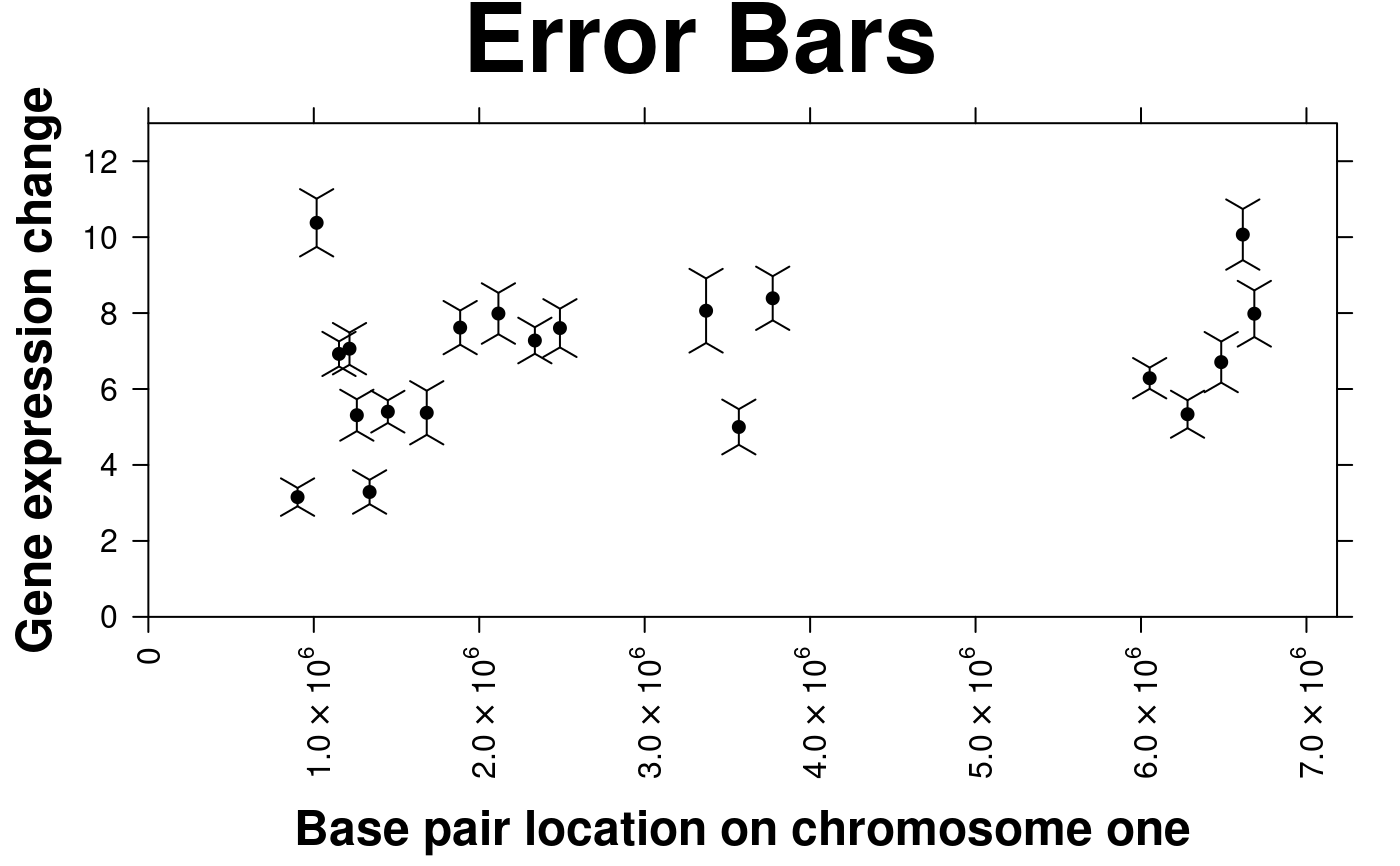
 # Error bars
error.data <- data.frame(
chr = (microarray$Start)[1:20],
values = apply(microarray[1:20,1:58], 1, mean),
error = apply(microarray[1:20,1:58], 1, sd)
);
create.scatterplot(
# filename = tempfile(pattern = 'Scatterplot_Error_Bars', fileext = '.tiff'),
formula = values ~ chr,
data = error.data,
main = 'Error Bars',
xlab.label = 'Base pair location on chromosome one',
ylab.label = 'Gene expression change',
# xat = seq(0, 16, 2),
yat = seq(0, 14, 2),
# xlimits = c(0, 15),
ylimits = c(0, 13),
# Format xaxes
xaxis.lab = c(
scientific.notation(0, 1),
scientific.notation(1000000, 1),
scientific.notation(2000000, 1),
scientific.notation(3000000, 1),
scientific.notation(4000000, 1),
scientific.notation(5000000, 1),
scientific.notation(6000000, 1),
scientific.notation(7000000, 1)
),
xaxis.rot = 90,
xaxis.cex = 1,
yaxis.cex = 1,
xaxis.fontface = 1,
yaxis.fontface = 1,
xlab.cex = 1.5,
ylab.cex = 1.5,
pch = 19,
col = 'black',
fill = 'transparent',
# Specifying error bars
error.bar.lwd = 1,
error.whisker.angle = 120,
y.error.up = error.data$error,
y.error.bar.col = 'black',
description = 'Scatter plot created by BoutrosLab.plotting.general',
resolution = 200
);
# Error bars
error.data <- data.frame(
chr = (microarray$Start)[1:20],
values = apply(microarray[1:20,1:58], 1, mean),
error = apply(microarray[1:20,1:58], 1, sd)
);
create.scatterplot(
# filename = tempfile(pattern = 'Scatterplot_Error_Bars', fileext = '.tiff'),
formula = values ~ chr,
data = error.data,
main = 'Error Bars',
xlab.label = 'Base pair location on chromosome one',
ylab.label = 'Gene expression change',
# xat = seq(0, 16, 2),
yat = seq(0, 14, 2),
# xlimits = c(0, 15),
ylimits = c(0, 13),
# Format xaxes
xaxis.lab = c(
scientific.notation(0, 1),
scientific.notation(1000000, 1),
scientific.notation(2000000, 1),
scientific.notation(3000000, 1),
scientific.notation(4000000, 1),
scientific.notation(5000000, 1),
scientific.notation(6000000, 1),
scientific.notation(7000000, 1)
),
xaxis.rot = 90,
xaxis.cex = 1,
yaxis.cex = 1,
xaxis.fontface = 1,
yaxis.fontface = 1,
xlab.cex = 1.5,
ylab.cex = 1.5,
pch = 19,
col = 'black',
fill = 'transparent',
# Specifying error bars
error.bar.lwd = 1,
error.whisker.angle = 120,
y.error.up = error.data$error,
y.error.bar.col = 'black',
description = 'Scatter plot created by BoutrosLab.plotting.general',
resolution = 200
);
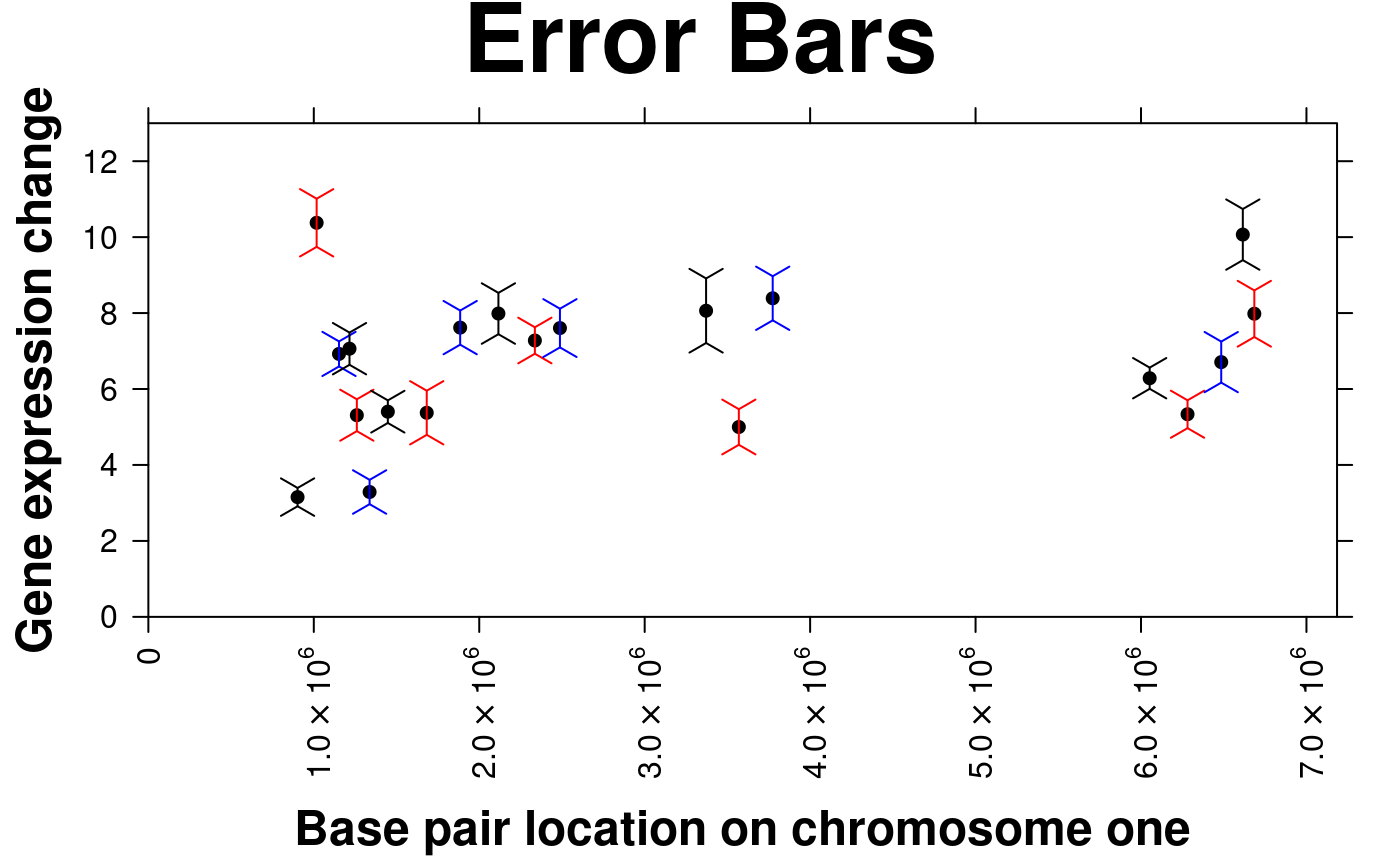
 create.scatterplot(
# filename = tempfile(pattern = 'Scatterplot_Error_Bars_MultiColor', fileext = '.tiff'),
formula = values ~ chr,
data = error.data,
main = 'Error Bars',
xlab.label = 'Base pair location on chromosome one',
ylab.label = 'Gene expression change',
# xat = seq(0, 16, 2),
yat = seq(0, 14, 2),
# xlimits = c(0, 15),
ylimits = c(0, 13),
# Format xaxes
xaxis.lab = c(
scientific.notation(0, 1),
scientific.notation(1000000, 1),
scientific.notation(2000000, 1),
scientific.notation(3000000, 1),
scientific.notation(4000000, 1),
scientific.notation(5000000, 1),
scientific.notation(6000000, 1),
scientific.notation(7000000, 1)
),
xaxis.rot = 90,
xaxis.cex = 1,
yaxis.cex = 1,
xaxis.fontface = 1,
yaxis.fontface = 1,
xlab.cex = 1.5,
ylab.cex = 1.5,
pch = 19,
col = 'black',
fill = 'transparent',
# Specifying error bars
error.bar.lwd = 1,
error.whisker.angle = 120,
y.error.up = error.data$error,
y.error.bar.col = c('black','red','blue'),
description = 'Scatter plot created by BoutrosLab.plotting.general',
group.specific.colouring = FALSE,
resolution = 200
);
create.scatterplot(
# filename = tempfile(pattern = 'Scatterplot_Error_Bars_MultiColor', fileext = '.tiff'),
formula = values ~ chr,
data = error.data,
main = 'Error Bars',
xlab.label = 'Base pair location on chromosome one',
ylab.label = 'Gene expression change',
# xat = seq(0, 16, 2),
yat = seq(0, 14, 2),
# xlimits = c(0, 15),
ylimits = c(0, 13),
# Format xaxes
xaxis.lab = c(
scientific.notation(0, 1),
scientific.notation(1000000, 1),
scientific.notation(2000000, 1),
scientific.notation(3000000, 1),
scientific.notation(4000000, 1),
scientific.notation(5000000, 1),
scientific.notation(6000000, 1),
scientific.notation(7000000, 1)
),
xaxis.rot = 90,
xaxis.cex = 1,
yaxis.cex = 1,
xaxis.fontface = 1,
yaxis.fontface = 1,
xlab.cex = 1.5,
ylab.cex = 1.5,
pch = 19,
col = 'black',
fill = 'transparent',
# Specifying error bars
error.bar.lwd = 1,
error.whisker.angle = 120,
y.error.up = error.data$error,
y.error.bar.col = c('black','red','blue'),
description = 'Scatter plot created by BoutrosLab.plotting.general',
group.specific.colouring = FALSE,
resolution = 200
);
 # Gridlines
create.scatterplot(
# filename = tempfile(pattern = 'Scatterplot_Gridlines', fileext = '.tiff'),
formula = sample.two ~ sample.one,
data = scatter.data,
main = 'Gridlines',
xlab.label = colnames(microarray[1]),
ylab.label = colnames(microarray[2]),
xat = seq(0, 16, 2),
yat = seq(0, 16, 2),
xlimits = c(0, 15),
ylimits = c(0, 15),
xaxis.cex = 1,
yaxis.cex = 1,
xaxis.fontface = 1,
yaxis.fontface = 1,
xlab.cex = 1.5,
ylab.cex = 1.5,
pch = 21,
col = 'black',
fill = 'transparent',
# Adding gridlines
type = c('p','g'),
description = 'Scatter plot created by BoutrosLab.plotting.general',
resolution = 200
);
# Gridlines
create.scatterplot(
# filename = tempfile(pattern = 'Scatterplot_Gridlines', fileext = '.tiff'),
formula = sample.two ~ sample.one,
data = scatter.data,
main = 'Gridlines',
xlab.label = colnames(microarray[1]),
ylab.label = colnames(microarray[2]),
xat = seq(0, 16, 2),
yat = seq(0, 16, 2),
xlimits = c(0, 15),
ylimits = c(0, 15),
xaxis.cex = 1,
yaxis.cex = 1,
xaxis.fontface = 1,
yaxis.fontface = 1,
xlab.cex = 1.5,
ylab.cex = 1.5,
pch = 21,
col = 'black',
fill = 'transparent',
# Adding gridlines
type = c('p','g'),
description = 'Scatter plot created by BoutrosLab.plotting.general',
resolution = 200
);
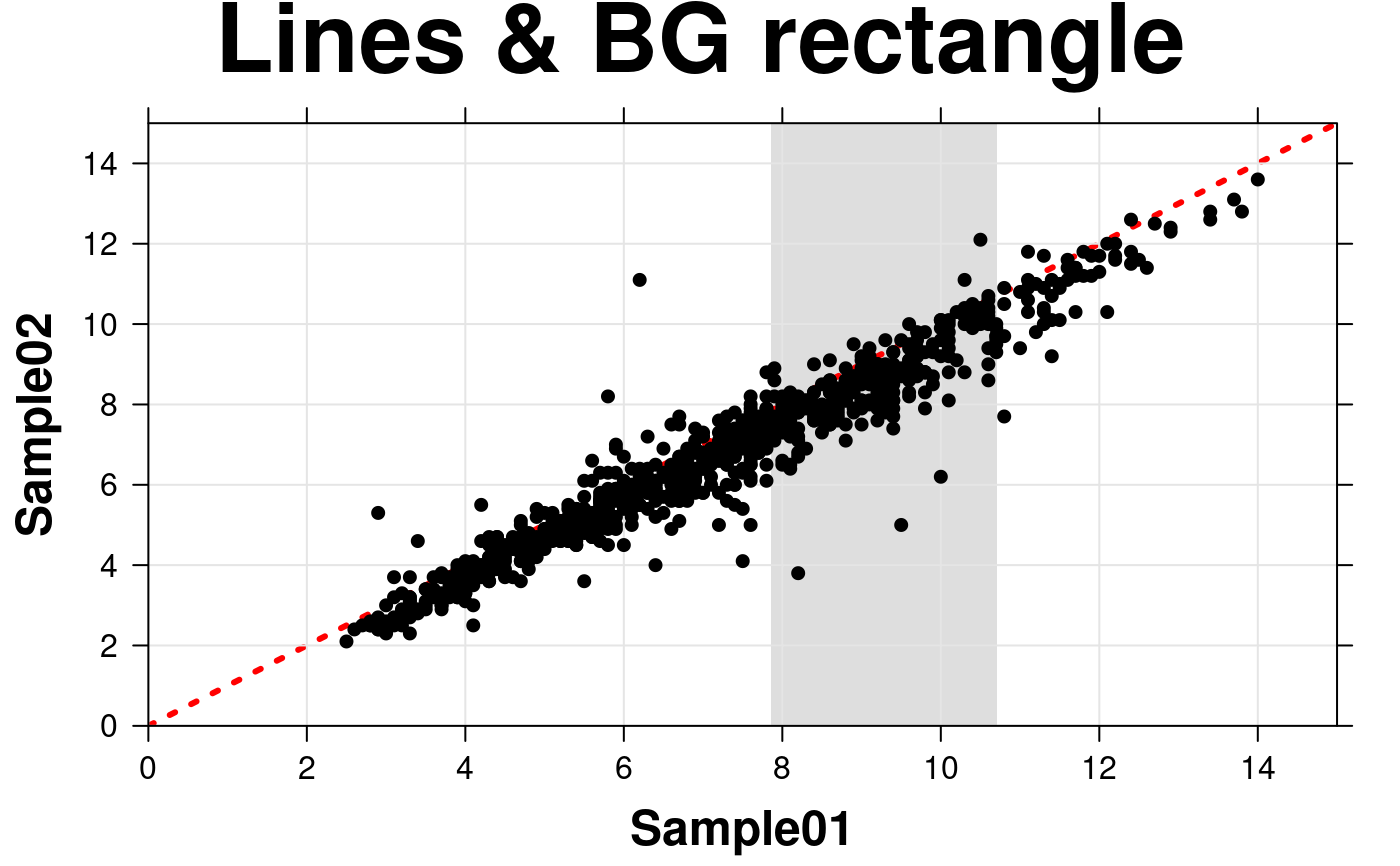
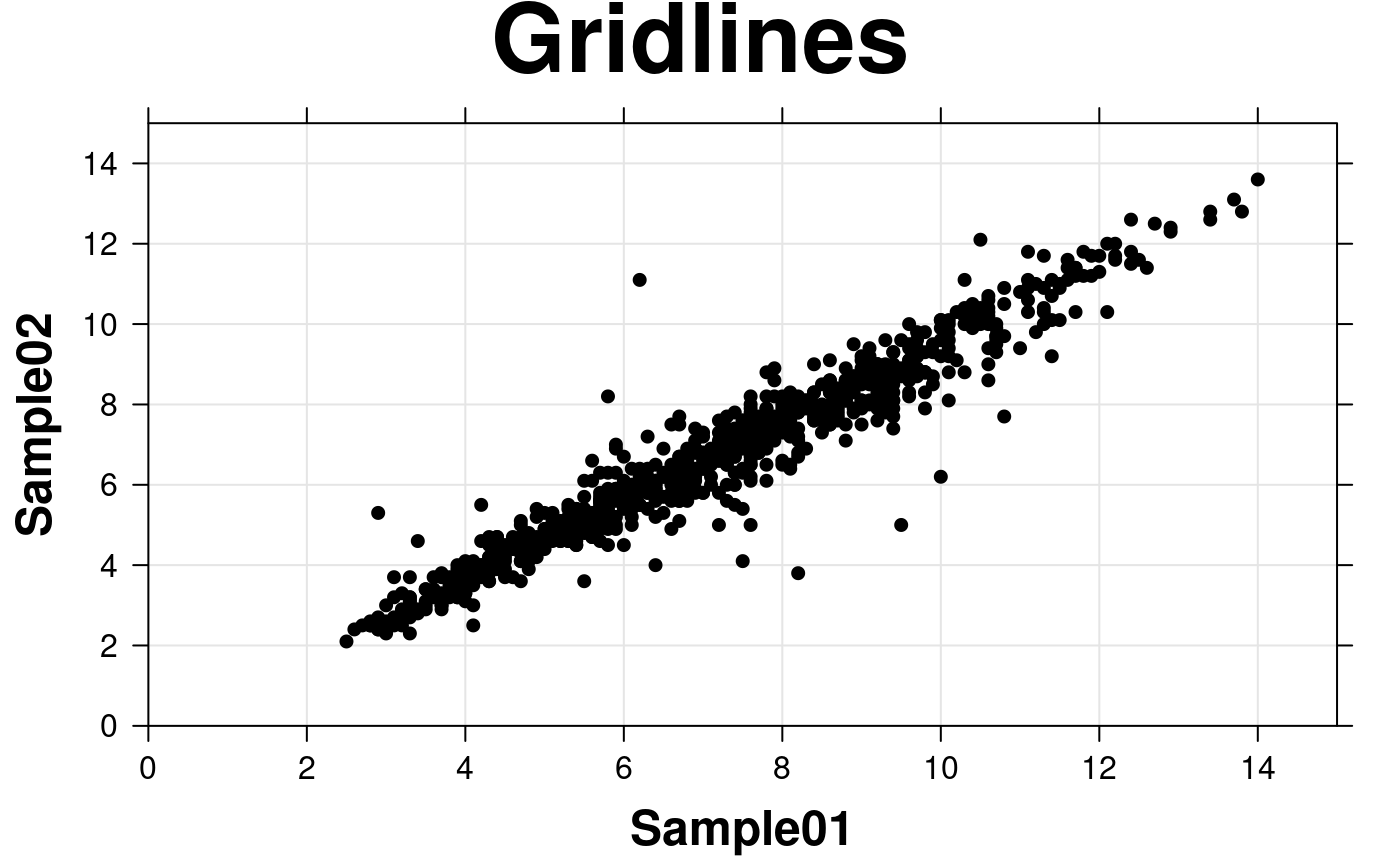 # lines & background rectangle
create.scatterplot(
# filename = tempfile(pattern = 'Scatterplot_Lines_BG', fileext = '.tiff'),
formula = sample.two ~ sample.one,
data = scatter.data,
main = 'Lines & BG rectangle',
xlab.label = colnames(microarray[1]),
ylab.label = colnames(microarray[2]),
xat = seq(0, 16, 2),
yat = seq(0, 16, 2),
xlimits = c(0, 15),
ylimits = c(0, 15),
xaxis.cex = 1,
yaxis.cex = 1,
xaxis.fontface = 1,
yaxis.fontface = 1,
xlab.cex = 1.5,
ylab.cex = 1.5,
pch = 21,
col = 'black',
fill = 'transparent',
type = c('p','g'),
# add xy line
add.xyline = TRUE,
xyline.lty = 3,
xyline.col = 'red',
xyline.lwd = 3,
# add background rectangle
add.rectangle = TRUE,
xleft.rectangle = which(scatter.data$chr == 2)[1]/800*15,
xright.rectangle = which(scatter.data$chr == 3)[1]/800*15,
ybottom.rectangle = 0,
ytop.rectangle = 15,
col.rectangle = 'grey',
alpha.rectangle = 0.5,
description = 'Scatter plot created by BoutrosLab.plotting.general',
resolution = 200
);
# lines & background rectangle
create.scatterplot(
# filename = tempfile(pattern = 'Scatterplot_Lines_BG', fileext = '.tiff'),
formula = sample.two ~ sample.one,
data = scatter.data,
main = 'Lines & BG rectangle',
xlab.label = colnames(microarray[1]),
ylab.label = colnames(microarray[2]),
xat = seq(0, 16, 2),
yat = seq(0, 16, 2),
xlimits = c(0, 15),
ylimits = c(0, 15),
xaxis.cex = 1,
yaxis.cex = 1,
xaxis.fontface = 1,
yaxis.fontface = 1,
xlab.cex = 1.5,
ylab.cex = 1.5,
pch = 21,
col = 'black',
fill = 'transparent',
type = c('p','g'),
# add xy line
add.xyline = TRUE,
xyline.lty = 3,
xyline.col = 'red',
xyline.lwd = 3,
# add background rectangle
add.rectangle = TRUE,
xleft.rectangle = which(scatter.data$chr == 2)[1]/800*15,
xright.rectangle = which(scatter.data$chr == 3)[1]/800*15,
ybottom.rectangle = 0,
ytop.rectangle = 15,
col.rectangle = 'grey',
alpha.rectangle = 0.5,
description = 'Scatter plot created by BoutrosLab.plotting.general',
resolution = 200
);
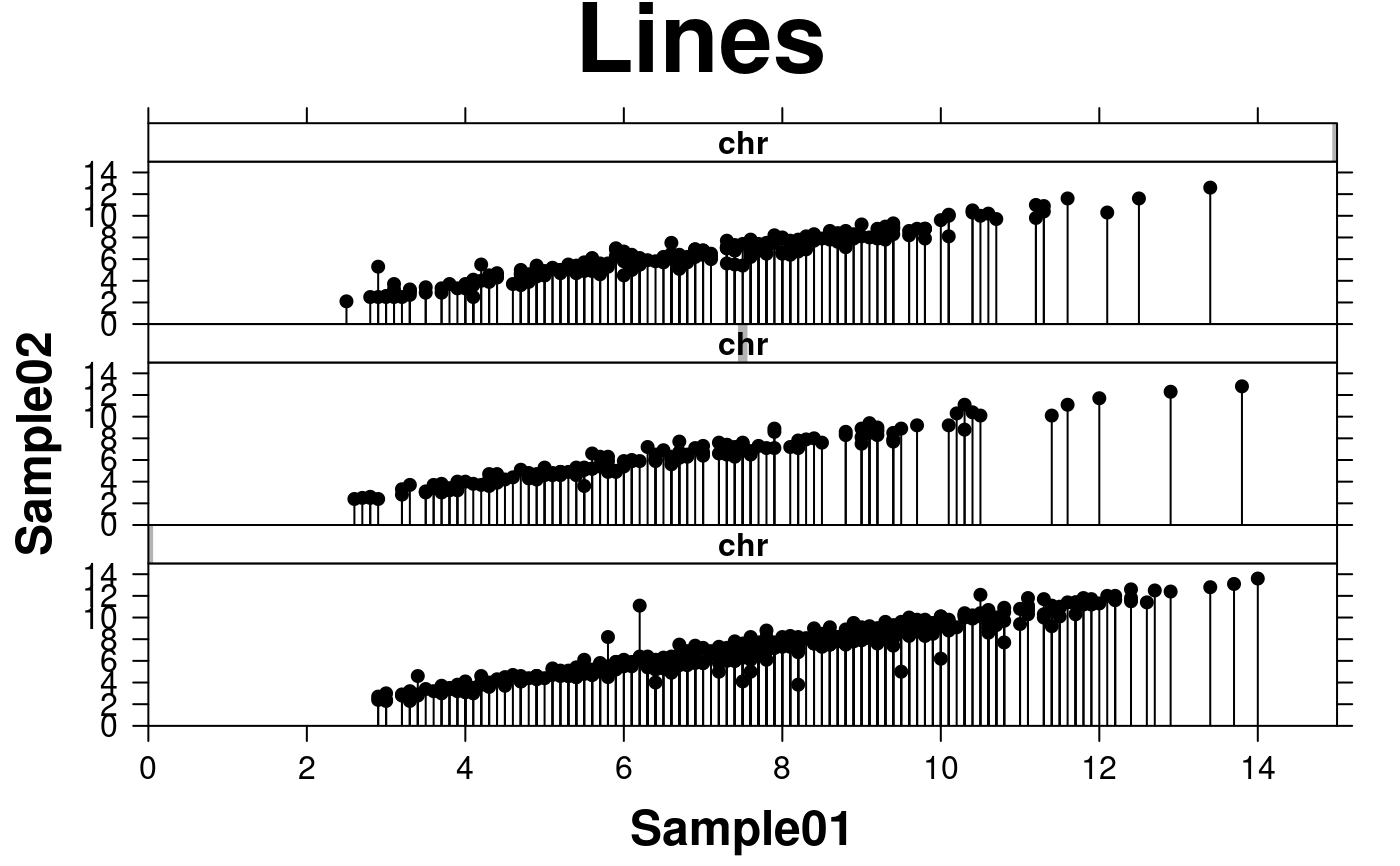
 # attach lines to points
create.scatterplot(
# filename = tempfile(pattern = 'Scatterplot_Lines', fileext = '.tiff'),
formula = sample.two ~ sample.one | chr,
data = scatter.data,
main = 'Lines',
xlab.label = colnames(microarray[1]),
ylab.label = colnames(microarray[2]),
xat = seq(0, 16, 2),
yat = seq(0, 16, 2),
xlimits = c(0, 15),
ylimits = c(0, 15),
xaxis.cex = 1,
yaxis.cex = 1,
xaxis.fontface = 1,
yaxis.fontface = 1,
xlab.cex = 1.5,
ylab.cex = 1.5,
pch = 21,
col = 'black',
fill = 'transparent',
# attach lines
type = c('h','p'),
layout = c(1,3),
description = 'Scatter plot created by BoutrosLab.plotting.general',
resolution = 200
);
#> Warning: Numeric values detected for conditional variable. If text labels are desired, please convert conditional variable to character.
# attach lines to points
create.scatterplot(
# filename = tempfile(pattern = 'Scatterplot_Lines', fileext = '.tiff'),
formula = sample.two ~ sample.one | chr,
data = scatter.data,
main = 'Lines',
xlab.label = colnames(microarray[1]),
ylab.label = colnames(microarray[2]),
xat = seq(0, 16, 2),
yat = seq(0, 16, 2),
xlimits = c(0, 15),
ylimits = c(0, 15),
xaxis.cex = 1,
yaxis.cex = 1,
xaxis.fontface = 1,
yaxis.fontface = 1,
xlab.cex = 1.5,
ylab.cex = 1.5,
pch = 21,
col = 'black',
fill = 'transparent',
# attach lines
type = c('h','p'),
layout = c(1,3),
description = 'Scatter plot created by BoutrosLab.plotting.general',
resolution = 200
);
#> Warning: Numeric values detected for conditional variable. If text labels are desired, please convert conditional variable to character.
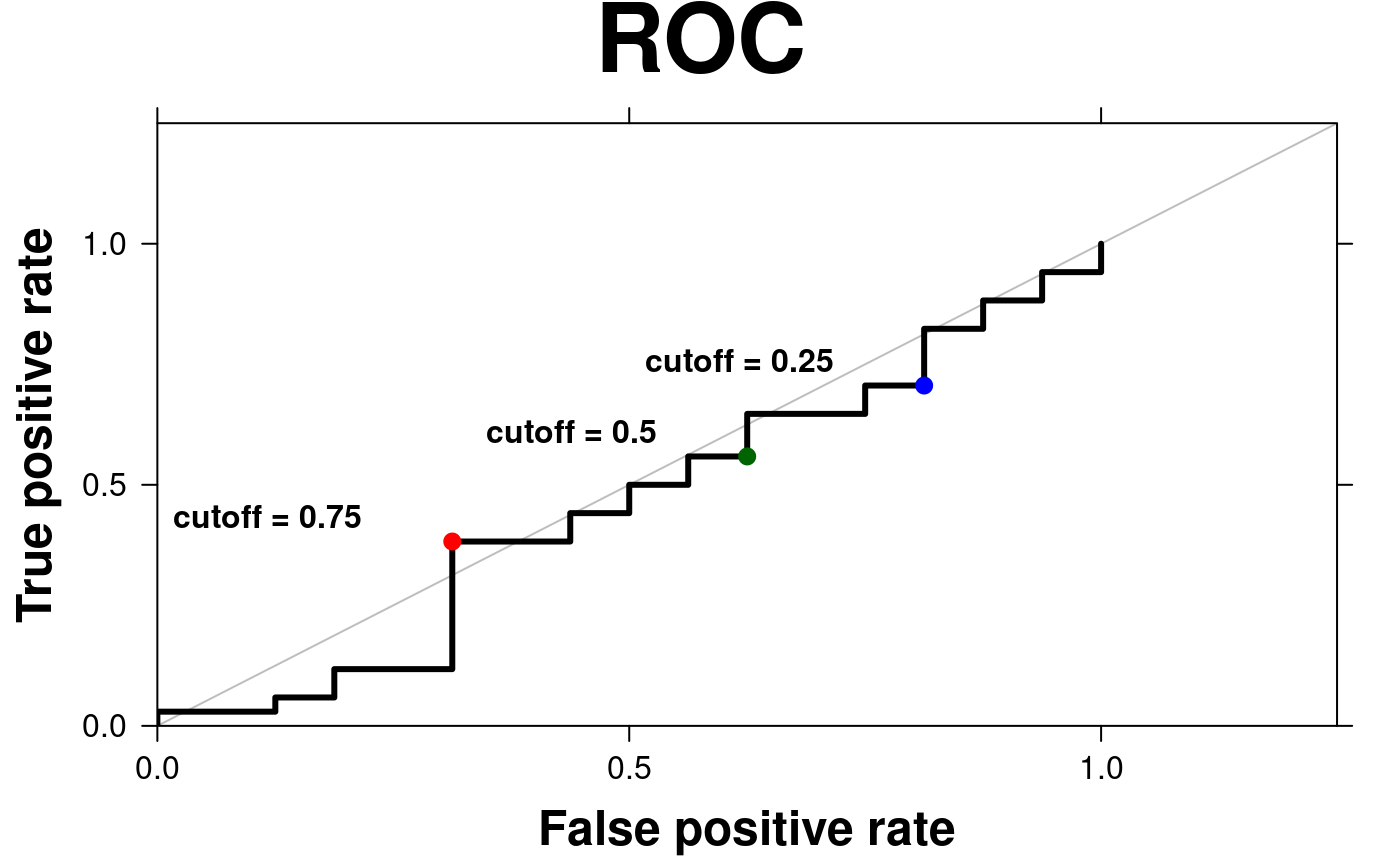
 # ROC curve
set.seed(123456);
class.values <- runif(50, 0, 1);
observed.values <- sample(c(0,1), size = 50, replace = TRUE);
cutoffs <- seq(1,0,-0.01);
tprs <- c();
fprs <- c();
for (c in cutoffs) {
roc.classification <- rep(0, length(class.values));
roc.classification[class.values >= c] <- 1;
roc.results <- table(
factor(roc.classification, levels = c(0,1)),
factor(observed.values, levels = c(0,1)),
dnn = c('pred', 'obs')
);
tprs <- c(tprs, roc.results[2,2] / (roc.results[2,2] + roc.results[1,2]));
fprs <- c(fprs, roc.results[2,1] / (roc.results[2,1] + roc.results[1,1]));
}
roc.data <- data.frame(cutoff = cutoffs, TPR = tprs, FPR = fprs);
points.x <- roc.data[match(c(0.25, 0.5, 0.75), roc.data$cutoff), 'FPR'];
points.y <- roc.data[match(c(0.25, 0.5, 0.75), roc.data$cutoff), 'TPR'];
create.scatterplot(
# filename = tempfile(pattern = 'Scatterplot_ROC', fileext = '.tiff'),
formula = TPR ~ FPR,
data = roc.data,
main = 'ROC',
xlab.label = 'False positive rate',
ylab.label = 'True positive rate',
xaxis.cex = 1,
yaxis.cex = 1,
xaxis.fontface = 1,
yaxis.fontface = 1,
xlab.cex = 1.5,
ylab.cex = 1.5,
# To plot ROC curve, add "s" or "S" to type vector.
# "s" connects points with the vertical segment first.
# "S" connects points with the horizontal segment first.
type = 's',
lwd = 3,
add.xyline = TRUE,
xyline.col = 'grey',
add.points = TRUE,
points.x = points.x,
points.y = points.y,
points.col = c('blue', 'darkgreen', 'red'),
add.text = TRUE,
text.labels = paste('cutoff = ', c(0.25, 0.5, 0.75), sep = ''),
#text.x = points.x - 0.14,
#text.y = points.y + 0.03,
text.x = points.x,
text.y = points.y,
text.guess.labels = TRUE,
text.guess.label.position = 155,
text.guess.radius.factor = 2.5,
description = 'Scatter plot created by BoutrosLab.plotting.general',
resolution = 200
);
# ROC curve
set.seed(123456);
class.values <- runif(50, 0, 1);
observed.values <- sample(c(0,1), size = 50, replace = TRUE);
cutoffs <- seq(1,0,-0.01);
tprs <- c();
fprs <- c();
for (c in cutoffs) {
roc.classification <- rep(0, length(class.values));
roc.classification[class.values >= c] <- 1;
roc.results <- table(
factor(roc.classification, levels = c(0,1)),
factor(observed.values, levels = c(0,1)),
dnn = c('pred', 'obs')
);
tprs <- c(tprs, roc.results[2,2] / (roc.results[2,2] + roc.results[1,2]));
fprs <- c(fprs, roc.results[2,1] / (roc.results[2,1] + roc.results[1,1]));
}
roc.data <- data.frame(cutoff = cutoffs, TPR = tprs, FPR = fprs);
points.x <- roc.data[match(c(0.25, 0.5, 0.75), roc.data$cutoff), 'FPR'];
points.y <- roc.data[match(c(0.25, 0.5, 0.75), roc.data$cutoff), 'TPR'];
create.scatterplot(
# filename = tempfile(pattern = 'Scatterplot_ROC', fileext = '.tiff'),
formula = TPR ~ FPR,
data = roc.data,
main = 'ROC',
xlab.label = 'False positive rate',
ylab.label = 'True positive rate',
xaxis.cex = 1,
yaxis.cex = 1,
xaxis.fontface = 1,
yaxis.fontface = 1,
xlab.cex = 1.5,
ylab.cex = 1.5,
# To plot ROC curve, add "s" or "S" to type vector.
# "s" connects points with the vertical segment first.
# "S" connects points with the horizontal segment first.
type = 's',
lwd = 3,
add.xyline = TRUE,
xyline.col = 'grey',
add.points = TRUE,
points.x = points.x,
points.y = points.y,
points.col = c('blue', 'darkgreen', 'red'),
add.text = TRUE,
text.labels = paste('cutoff = ', c(0.25, 0.5, 0.75), sep = ''),
#text.x = points.x - 0.14,
#text.y = points.y + 0.03,
text.x = points.x,
text.y = points.y,
text.guess.labels = TRUE,
text.guess.label.position = 155,
text.guess.radius.factor = 2.5,
description = 'Scatter plot created by BoutrosLab.plotting.general',
resolution = 200
);
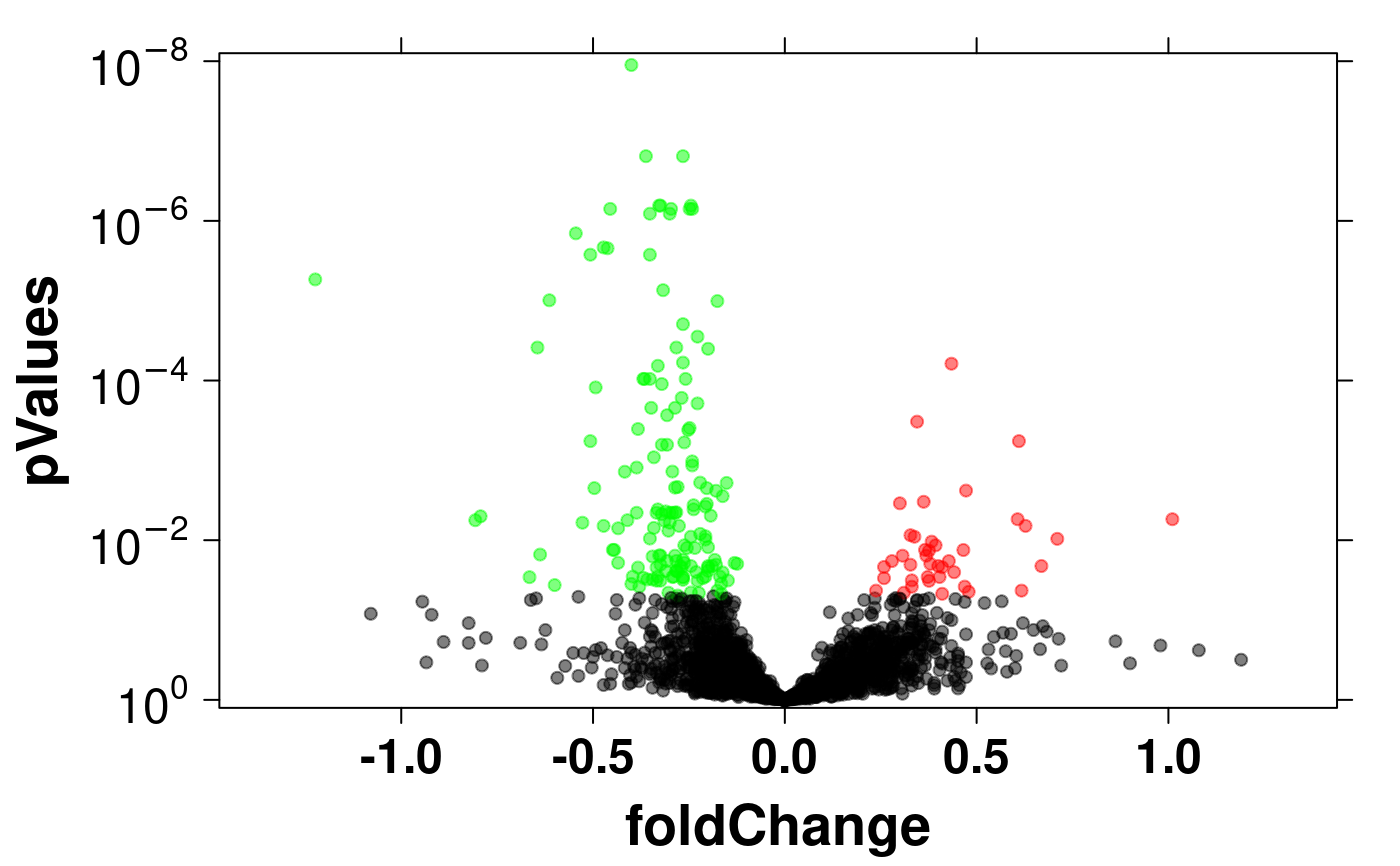
 # Volcano Plots
fold.change <- apply(microarray[,1:29], 1, mean) - apply(microarray[,30:58], 1, mean);
fake.microarray <- microarray[,1:58] - log(mean(apply(microarray[,1:58],1, mean)));
fake.microarray[,30:58] <- fake.microarray[,30:58] + mean(fold.change);
fake.microarray[fake.microarray < 0] <- 0;
p.values <- apply(fake.microarray[,1:58], 1, function(x) {t.test(x=x[1:29],y=x[30:58])$p.value} );
fold.change <- apply(fake.microarray[, 1:29], 1, mean) - apply(fake.microarray[, 30:58], 1,mean);
p.values.adjusted <- p.adjust(p.values, 'fdr');
dot.colours <- vector(length=length(p.values));
dot.colours[p.values.adjusted < .05 & fold.change < 0] <- 'green';
dot.colours[p.values.adjusted < .05 & fold.change > 0] <- 'red';
dot.colours[p.values.adjusted > .05] <- 'black';
volcano.data <- data.frame(
p.values = -log10(p.values.adjusted),
fold.change = fold.change
);
create.scatterplot(
# filename = tempfile(pattern = 'Scatterplot_Volcano_Plot', fileext = '.tiff'),
formula = p.values ~ fold.change,
data = volcano.data,
col = dot.colours,
alpha = .5,
yat = c(0,2,4,6,8),
ylimits = c(-0.1,8.1),
yaxis.lab = expression(10^0,10^-2,10^-4,10^-6,10^-8),
yaxis.cex = 1.5,
xaxis.cex = 1.5,
xlab.label = 'foldChange',
ylab.label = 'pValues',
xlab.cex = 1.75,
ylab.cex = 1.75,
resolution = 200
);
# Volcano Plots
fold.change <- apply(microarray[,1:29], 1, mean) - apply(microarray[,30:58], 1, mean);
fake.microarray <- microarray[,1:58] - log(mean(apply(microarray[,1:58],1, mean)));
fake.microarray[,30:58] <- fake.microarray[,30:58] + mean(fold.change);
fake.microarray[fake.microarray < 0] <- 0;
p.values <- apply(fake.microarray[,1:58], 1, function(x) {t.test(x=x[1:29],y=x[30:58])$p.value} );
fold.change <- apply(fake.microarray[, 1:29], 1, mean) - apply(fake.microarray[, 30:58], 1,mean);
p.values.adjusted <- p.adjust(p.values, 'fdr');
dot.colours <- vector(length=length(p.values));
dot.colours[p.values.adjusted < .05 & fold.change < 0] <- 'green';
dot.colours[p.values.adjusted < .05 & fold.change > 0] <- 'red';
dot.colours[p.values.adjusted > .05] <- 'black';
volcano.data <- data.frame(
p.values = -log10(p.values.adjusted),
fold.change = fold.change
);
create.scatterplot(
# filename = tempfile(pattern = 'Scatterplot_Volcano_Plot', fileext = '.tiff'),
formula = p.values ~ fold.change,
data = volcano.data,
col = dot.colours,
alpha = .5,
yat = c(0,2,4,6,8),
ylimits = c(-0.1,8.1),
yaxis.lab = expression(10^0,10^-2,10^-4,10^-6,10^-8),
yaxis.cex = 1.5,
xaxis.cex = 1.5,
xlab.label = 'foldChange',
ylab.label = 'pValues',
xlab.cex = 1.75,
ylab.cex = 1.75,
resolution = 200
);
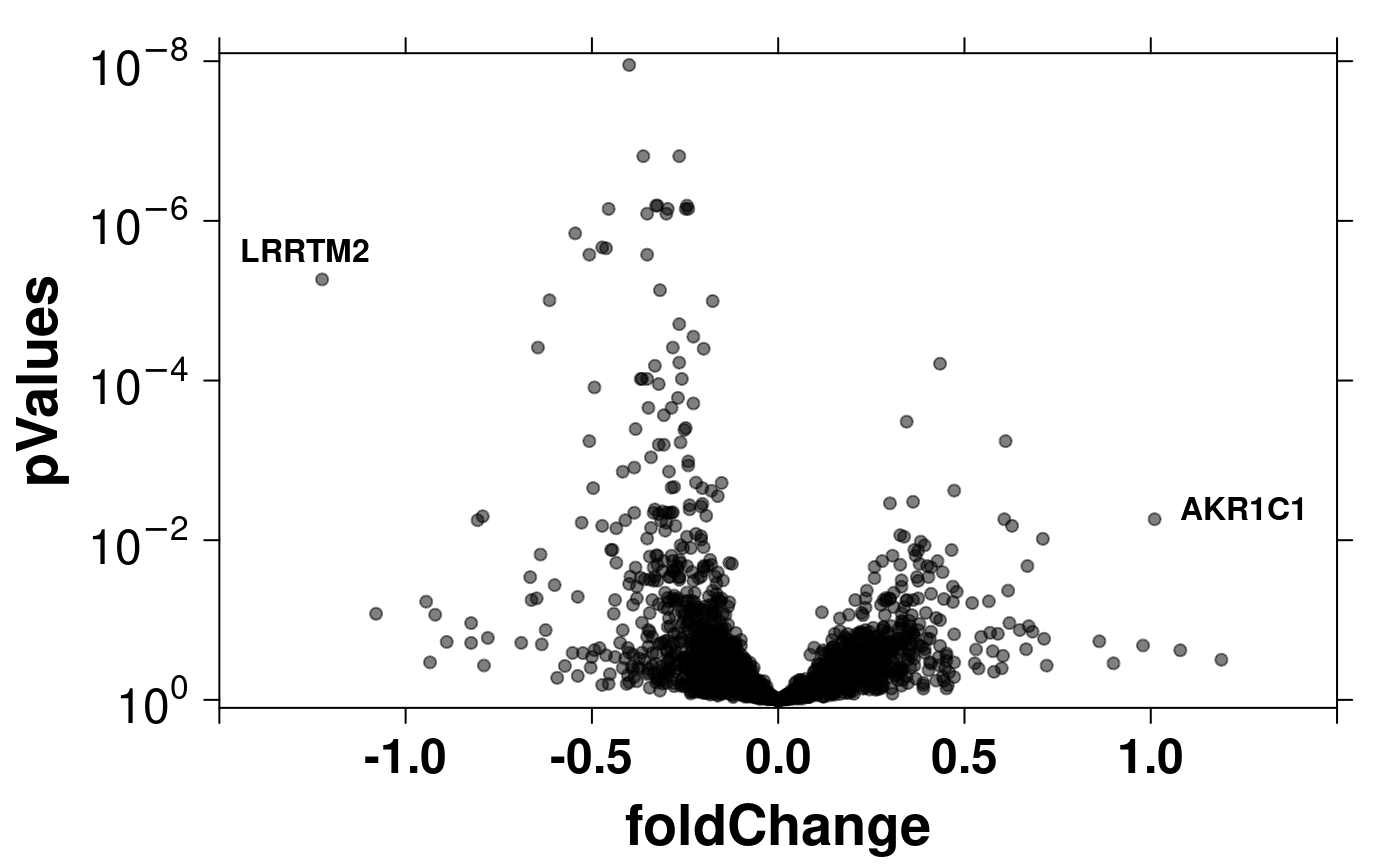
 # Automatic Labeling
interesting.fold.change <- (fold.change < -.9 | fold.change > .9);
interesting.p.value <- (-log10(p.values.adjusted) < 8 & -log10(p.values.adjusted) > 2);
interesting.points <- interesting.fold.change & interesting.p.value;
text.x <- fold.change[interesting.points];
text.y <- (-log10(p.values.adjusted))[interesting.points];
text.labels <- rownames(microarray)[interesting.points];
create.scatterplot(
# filename = tempfile(pattern = 'Scatterplot_Volcano_Plot_With_Labels', fileext = '.tiff'),
formula = p.values ~ fold.change,
data = volcano.data,
alpha = .5,
yat = c(0,2,4,6,8),
ylimits = c(-0.1,8.1),
xlimits = c(-1.5,1.5),
yaxis.lab = expression(10^0,10^-2,10^-4,10^-6,10^-8),
yaxis.cex = 1.5,
xaxis.cex = 1.5,
xlab.label = 'foldChange',
ylab.label = 'pValues',
xlab.cex = 1.75,
ylab.cex = 1.75,
add.text = TRUE,
text.x = text.x,
text.y = text.y,
text.labels = text.labels,
text.guess.labels = TRUE,
resolution = 200
);
# Automatic Labeling
interesting.fold.change <- (fold.change < -.9 | fold.change > .9);
interesting.p.value <- (-log10(p.values.adjusted) < 8 & -log10(p.values.adjusted) > 2);
interesting.points <- interesting.fold.change & interesting.p.value;
text.x <- fold.change[interesting.points];
text.y <- (-log10(p.values.adjusted))[interesting.points];
text.labels <- rownames(microarray)[interesting.points];
create.scatterplot(
# filename = tempfile(pattern = 'Scatterplot_Volcano_Plot_With_Labels', fileext = '.tiff'),
formula = p.values ~ fold.change,
data = volcano.data,
alpha = .5,
yat = c(0,2,4,6,8),
ylimits = c(-0.1,8.1),
xlimits = c(-1.5,1.5),
yaxis.lab = expression(10^0,10^-2,10^-4,10^-6,10^-8),
yaxis.cex = 1.5,
xaxis.cex = 1.5,
xlab.label = 'foldChange',
ylab.label = 'pValues',
xlab.cex = 1.75,
ylab.cex = 1.75,
add.text = TRUE,
text.x = text.x,
text.y = text.y,
text.labels = text.labels,
text.guess.labels = TRUE,
resolution = 200
);
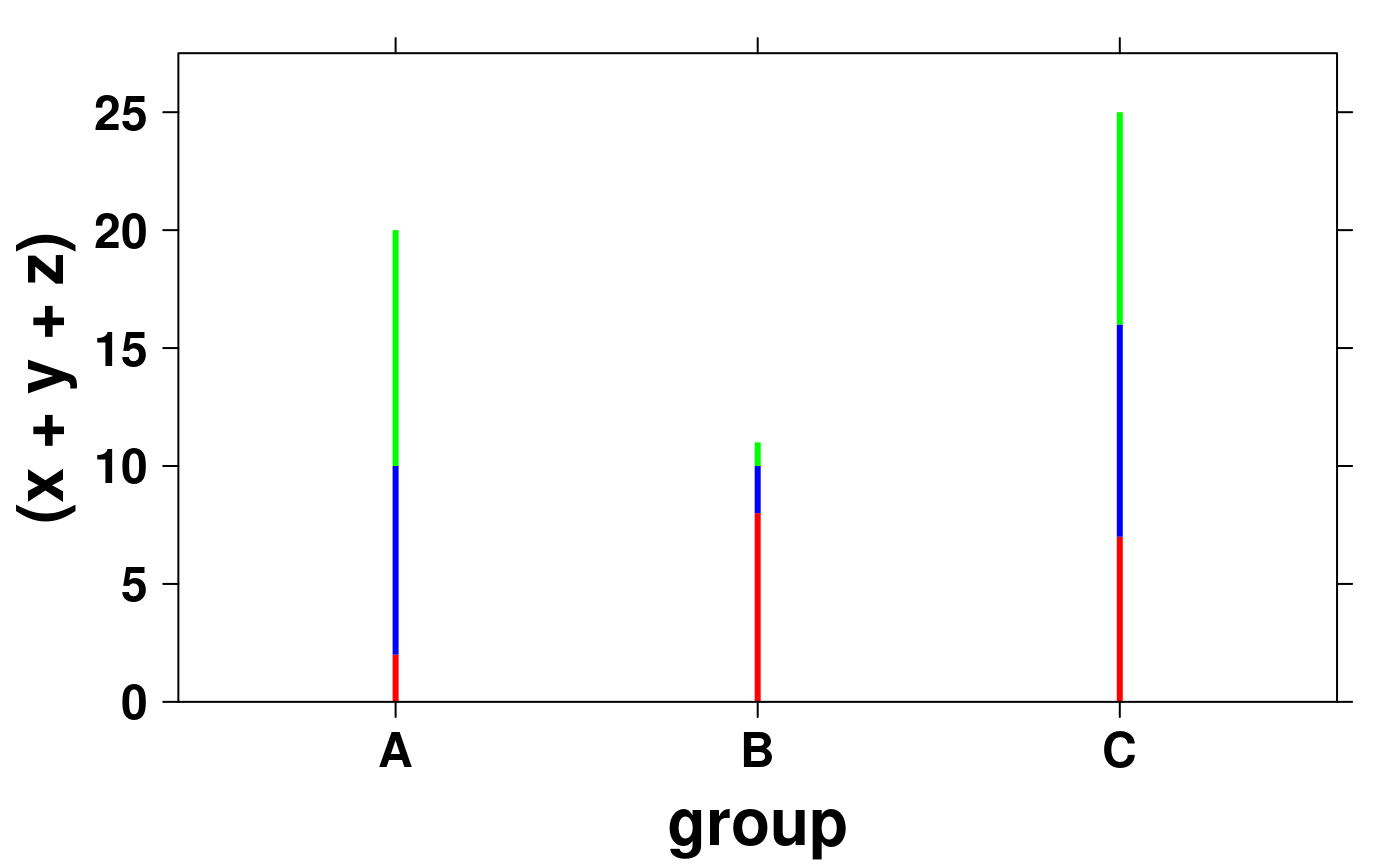
 # With line segments
line.data <- data.frame(
group = as.factor(c('A','B','C')),
x = sample(1:10,3),
y = sample(1:10,3),
z = sample(1:10,3)
);
create.scatterplot(
(x+y+z) ~ group,
line.data,
# filename = tempfile(pattern = 'Scatterplot_with_LineSegments', fileext = '.tiff'),
cex = 0,
add.line.segments = TRUE,
line.start = list(
rep(0,nrow(line.data)),
line.data$x,
c(line.data$x + line.data$y)
),
line.end = list(
line.data$x,
c(line.data$x + line.data$y),
c(line.data$x + line.data$y + line.data$z)
),
line.col = list('red','blue','green'),
line.lwd = list(3,3,3),
resolution = 200
);
# With line segments
line.data <- data.frame(
group = as.factor(c('A','B','C')),
x = sample(1:10,3),
y = sample(1:10,3),
z = sample(1:10,3)
);
create.scatterplot(
(x+y+z) ~ group,
line.data,
# filename = tempfile(pattern = 'Scatterplot_with_LineSegments', fileext = '.tiff'),
cex = 0,
add.line.segments = TRUE,
line.start = list(
rep(0,nrow(line.data)),
line.data$x,
c(line.data$x + line.data$y)
),
line.end = list(
line.data$x,
c(line.data$x + line.data$y),
c(line.data$x + line.data$y + line.data$z)
),
line.col = list('red','blue','green'),
line.lwd = list(3,3,3),
resolution = 200
);
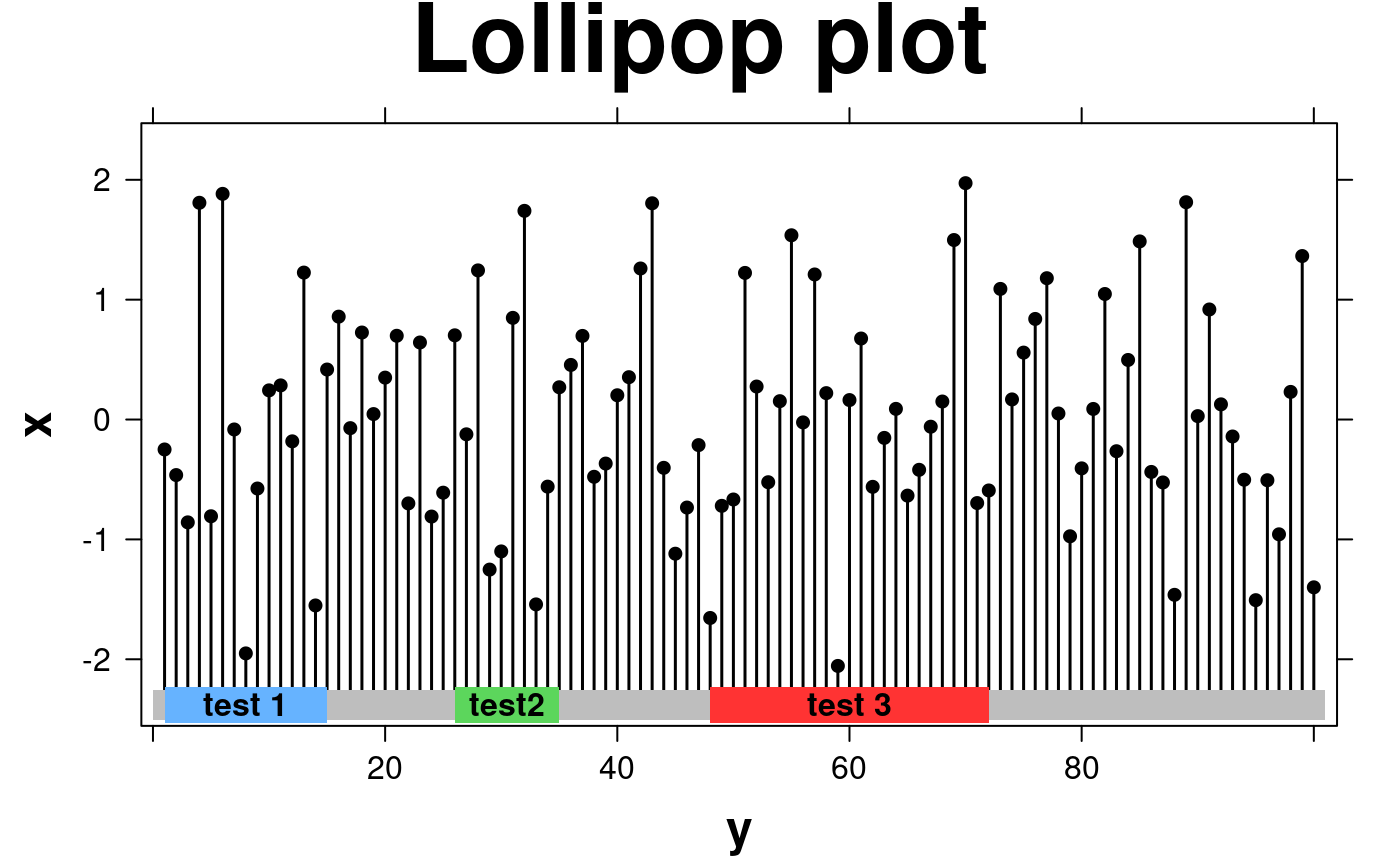
 lollipop.data <- data.frame(
y = seq(1,100,1),
x = rnorm(100)
);
create.lollipopplot(
# filename = tempfile(pattern = 'Lollipop_Simple', fileext = '.tiff'),
formula = x ~ y,
data = lollipop.data,
main = 'Lollipop plot',
xaxis.cex = 1,
xlimits = c(-1,102),
yaxis.cex = 1,
xaxis.fontface = 1,
yaxis.fontface = 1,
xlab.cex = 1.5,
ylab.cex = 1.5,
pch = 21,
col = 'black',
fill = 'transparent',
description = 'Scatter plot created by BoutrosLab.plotting.general',
regions.start = c(1,26,48),
regions.stop = c(15,35,72),
regions.labels = c("test 1", "test2", "test 3"),
regions.color = c("#66b3ff", "#5cd65c", "#ff3333"),
resolution = 200
);
lollipop.data <- data.frame(
y = seq(1,100,1),
x = rnorm(100)
);
create.lollipopplot(
# filename = tempfile(pattern = 'Lollipop_Simple', fileext = '.tiff'),
formula = x ~ y,
data = lollipop.data,
main = 'Lollipop plot',
xaxis.cex = 1,
xlimits = c(-1,102),
yaxis.cex = 1,
xaxis.fontface = 1,
yaxis.fontface = 1,
xlab.cex = 1.5,
ylab.cex = 1.5,
pch = 21,
col = 'black',
fill = 'transparent',
description = 'Scatter plot created by BoutrosLab.plotting.general',
regions.start = c(1,26,48),
regions.stop = c(15,35,72),
regions.labels = c("test 1", "test2", "test 3"),
regions.color = c("#66b3ff", "#5cd65c", "#ff3333"),
resolution = 200
);
 # }
# }